Lecture 7 / Chapter 8: Genetics in Bacteria and Bacteriophages
1/71
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
72 Terms
What is the name of the liquid upon which bacteria are grown?
Agar.
Colony?
A visible cluster of identical bacteria grown from an isolated bacterium on an agar plate.
Name two things that the study of bacteria is fundamental to:
CRISP-Cas (Recombinant DNA technology)
Insulin production (Medicine)
What unit is typically used when measuring the concentration of bacteria in a solution?
#cells/mL
What is the formula for bacteria in an original sample after serial dilutions?
original sample = colonies on a plate * dilution factor
Dilution factor is usually a multiple of 10.
Minimal media?
Media with the minimum amount of nutrients needed to live
What is contained within minimal media?
The minimum amount of nutrients for an organism to live, as the name suggests.
To be more specific, it contains an organic carbon source and a variety of inorganic ions.
There are two important phenotypes of bacteria that were considered during experiments involving horizontal gene transfer, and had to do with the bacteria’s ability to survive on minimal media.
Prototrophs and auxotrophs.
Prototroph?
Bacteria that can grow on minimal media, and are considered WT
proto = old, before
Auxotroph?
Cannot synthesize all essential organic compounds; cannot grow on minimal media.
Auxo = help
What are the two types of inheritance by direction?
Vertical gene transfer
Horizontal “ “
Vertical gene transfer?
Genetic information is transferred to offspring; transfer between organisms of the same species
Horizontal gene transfer?
Genetic information is transferred between organisms that are not parent and offpsirng
Is the transfer ABx resistance genes between bacteria an example of vertical or horizontal gene transfer?
HGT
The foundation for chromosome mapping in bacteria is
genetic recombination.
How does recombination take place in bacteria? How does this affect the bacteria? What is the result in different bacteria populations?
One or more genes present in a strain are replaced with genes from a genetically distinct strain.
The replacement of genes produces a new phenotype.
The result is genvar between strains.
What are the three modes of gene transfer in bacteria?
Conjugation
Transformation
Transduction
Conjugation?
Genetic material from one bacterium is transferred to another bacterium, and sometimes recombines
Describe the Set-up of the Lederberg-Tatum Experiment.
Involves two multiple-auxotrophs of E. coli strain K12.
One auxotroph was unable to produce met and bio.
The other auxotroph was unable to produce thr, leu, and thi.
The two strains were grown separately in supplemented media.
After being grown separately, they were mixed together and grown together for several generations.
The mixed bacteria were then introduced to minimal media.
Describe the results of the Lederberg-Tatum Experiment.
The procedure produced prototroph colonies that survived off minimal media.
Why can’t the presence of prototrophs in the minimal media be explained as being a result of mutation?
There were two or three mutant genes depending on the strain. It’s highly unlikely that a significant amount of these cells underwent spontaneous mutation at the exact location of these genes to produce a prototroph.
What conclusion did the Lederberg-Tatum Experiment come up with?
Any prototrophs that have arisen from the experiment occurred as a result of some form of genetic exchange and recombination between two mutant strains.
Describe the Set-up of the Davis U-tube Experiment.
Two auxotrophs are grown in a common medium (stored in a U-Tube) but separated by a filter.
Both sides have auxotrophs in a supplemented medium. What one side can produce, the other can’t.
Samples of either side are taken before and after the filter is removed to see if the bacteria can live on minimal media.
Describe the results of the Davis U-tube Experiment.
Before the filter is removed, bacteria from neither side could live on minimal media.
After the filter is removed, bacteria from both sides could live on minimal media.
What conclusion did the Davis U-tube Experiment come up with?
Physical contact is required for recombination
In conjugation, a ______ flow of information is observed
unidirectional
F factor?
A plasmid in bacterial cells that confers the ability to act as a donor in conjugation.
Donor cells are ____; Recipient cells are ____.
F+, F-
Compare the results of conjugation between F+ and F- vs. between Hfr and F-.
F+ x F- → Both donor and recipient end up F+; very little recombination occurs.
Hfr x F- → Donor and recipient still stay F+ and F- respectively. High levels of recombination, though.
Explain why the results of conjugation between F+ and F- vs. between Hfr and F- are different.
In both interactions, different things are being transferred over.
In F+ and F- conjugation, a copy of the F plasmid is given to the recipient, while chromosomal DNA usually is not.
In Hfr and F- conjugation, chromosomal DNA is replicated and given to the recipient, starting at the origin of transfer (O) but stopping before the F factor is reached.
How is an F+ cell converted into an Hfr cell?
Random integration of the F factor into the chromosome.
Hfr cells are a kind of _____ cell.
F+
What is the name of the process used to map genes in bacteria?
Interrupted mating technique
What are the steps to interrupted mating technique (IMT)?
Culture an Hfr strain with an F- strain.
Remove samples in intervals as culture grows.
Samples are placed in a blender to separate conjugating cells, stopping DNA transfer.
Determine the amount of DNA transferred to F- cells.
In IMT, why do we need to use Hfr strains instead of normal F+ strains?
In order to ‘see’ the gene, we need to see the phenotype in the recipient cell. Changes in phenotype occur due to recombination, which can occur when donor chromosomal DNA is given to a recipient cell and integrated into the recipient’s chromosome.
F+ strains do not donate chromosomal DNA, and instead only donate the F factor. Hfr strains do donate chromosomal DNA, which is why they’re necessary for IMT.
How do we determine the amount of DNA transferred to F- cells?
We can determine the amount of DNA transferred by looking at the number of F- cells that show certain phenotypes. Their phenotypes will reveal that they express certain genes. Genes will only be expressed after being integrated, and the order that they become integrated is dependent on the sequence. So, by looking at the changes in the phenotype of the recipient DNA, we can tell how much DNA has been transferred.
What did IMT reveal about conjugation? (Conclusions?)
Certain genes were transferred before others.
The time between transfer of genes can be used as a measure of distance between genes.
Transfer can proceed in (both / clockwise / counterclockwise) direction(s) starting from the origin (O)
both
Transformation?
Mechanism for uptake of DNA from the surroundings into a bacterial cell.
In what kind of environment does transformation take place?
In harsh environments (GGT)
Competent Cells?
Cells that are able to take up and incorporate foreign DNA into their genome.
What are the two ways that a cell can become competent?
Natural Competence: A cell becomes competent in response to harshened conditions.
Artificial / Induced Competence: Through electrical pulses or chemical treatments, a cell in lab is made more permeable to DNA.
How does transformation help us map genes?
Small pieces of DNA (10-20kb) are taken up by bacteria through a receptor-mediated mechanism.
DNA may be large enough to contain several genes allowing for genetic mapping.
Genes that are close together on the chromosome are more likely to cotransform; these genes are thus linked.
What unit is a kb?
kilobase (1000 nitrogenous bases)
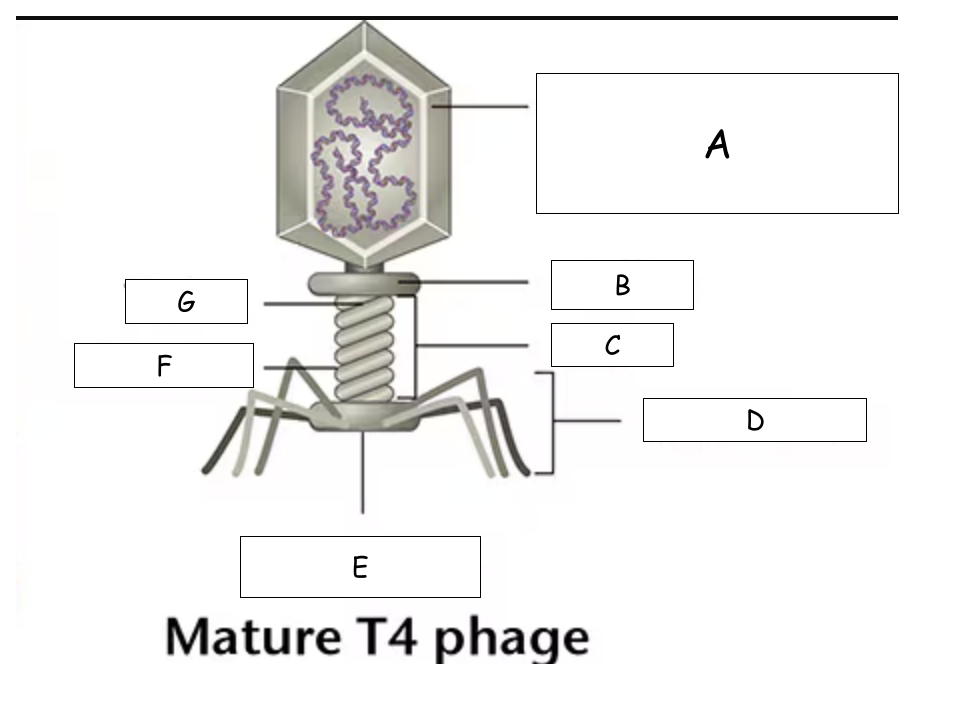
(See Mature T4 Phage.png) A?
Head (with packaged DNA)
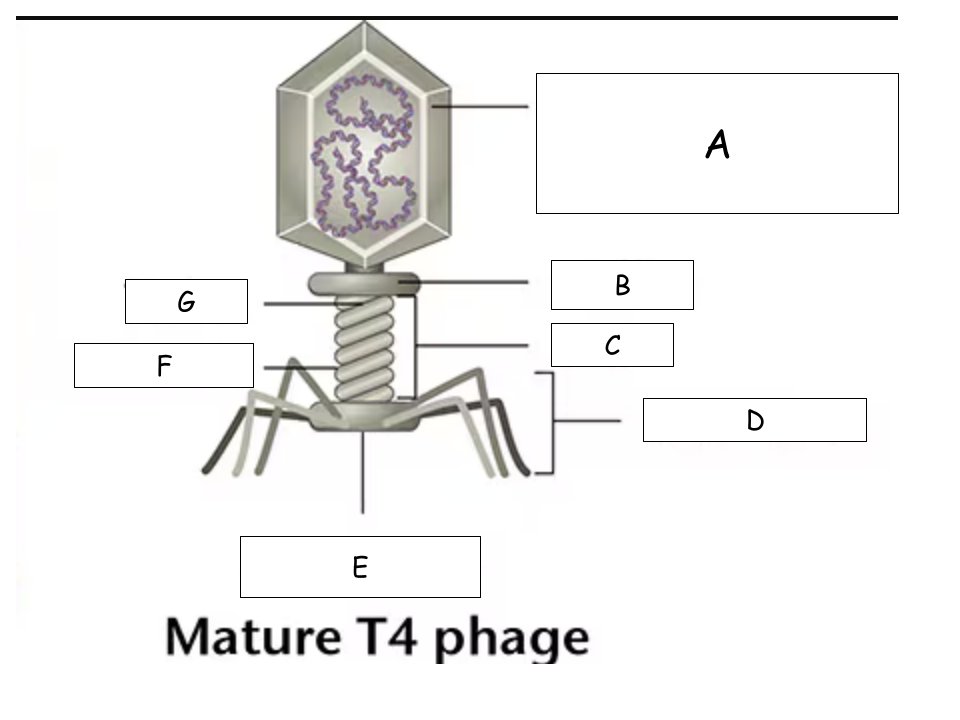
(See Mature T4 Phage.png) B?
Collar
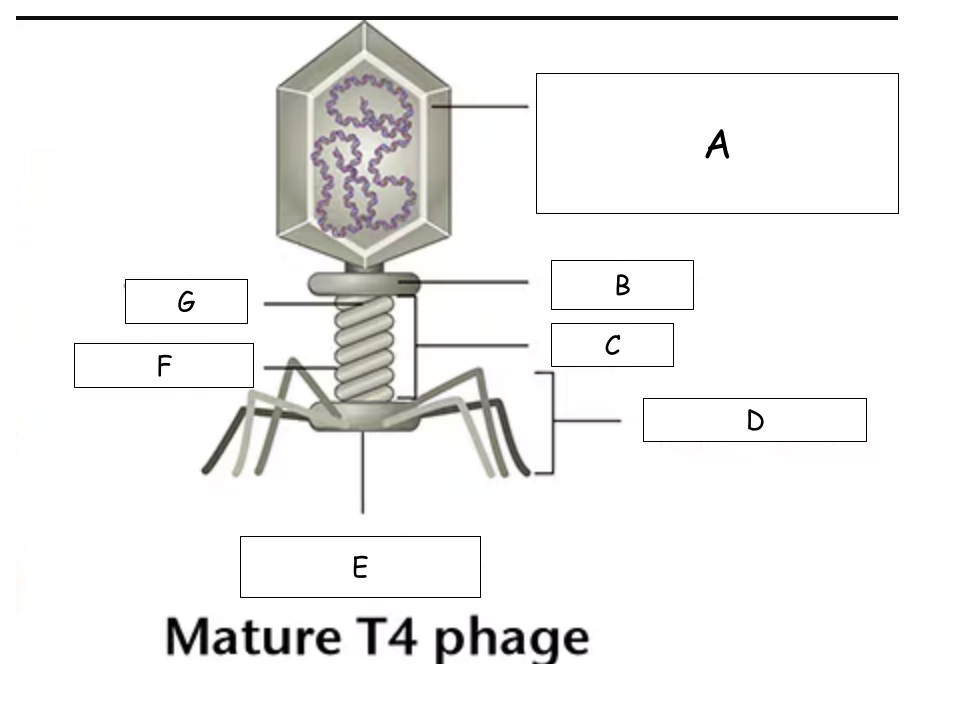
(See Mature T4 Phage.png) C?
Tail
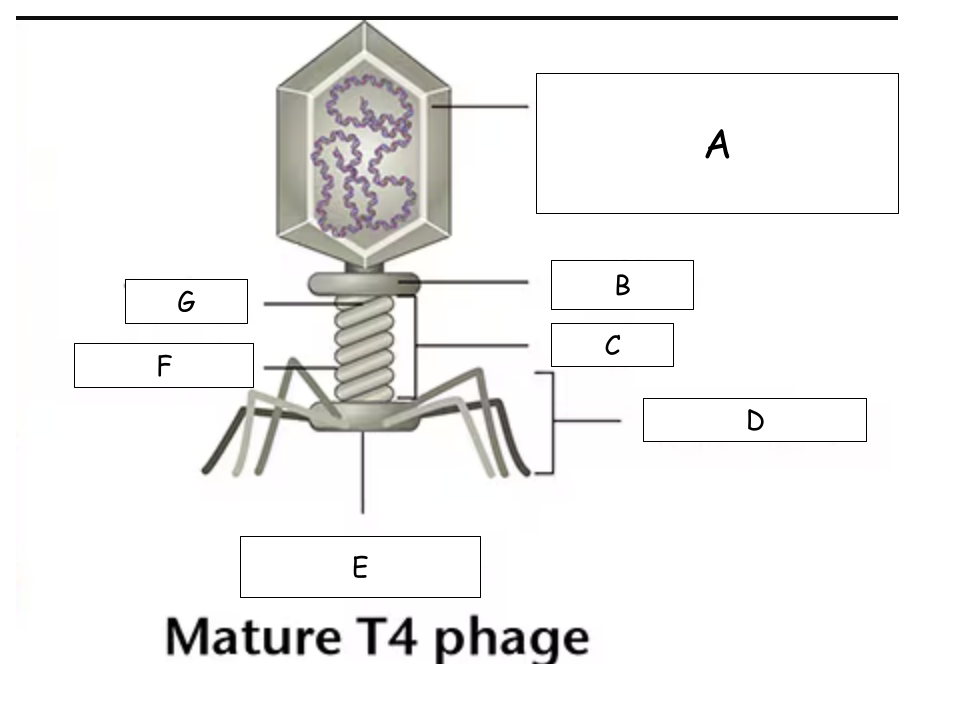
(See Mature T4 Phage.png) D?
Tail Fibers
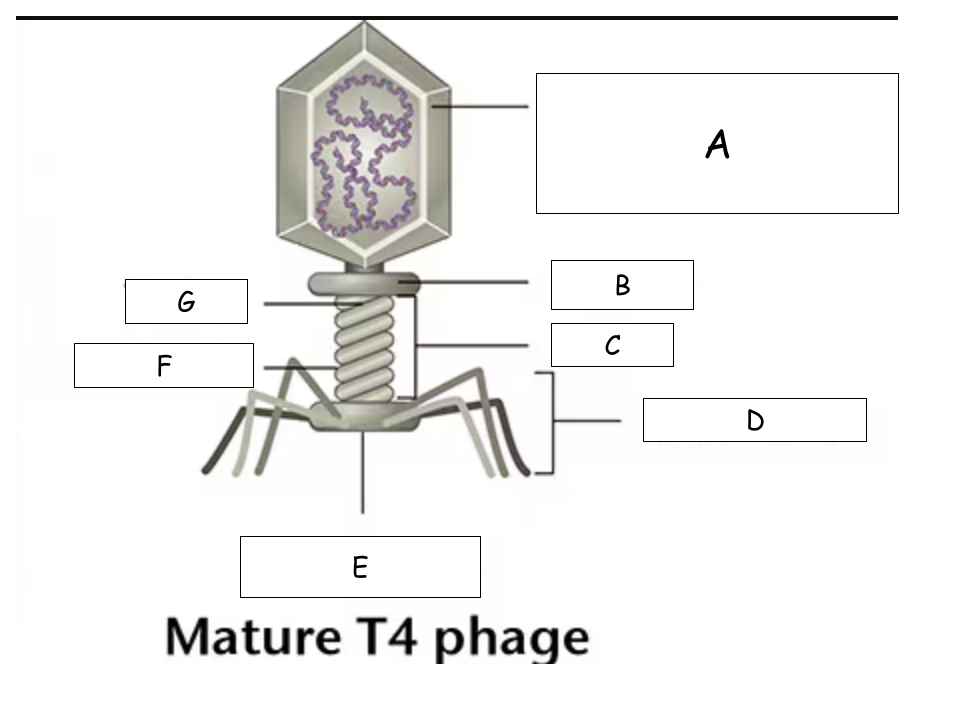
(See Mature T4 Phage.png) E?
Base plate
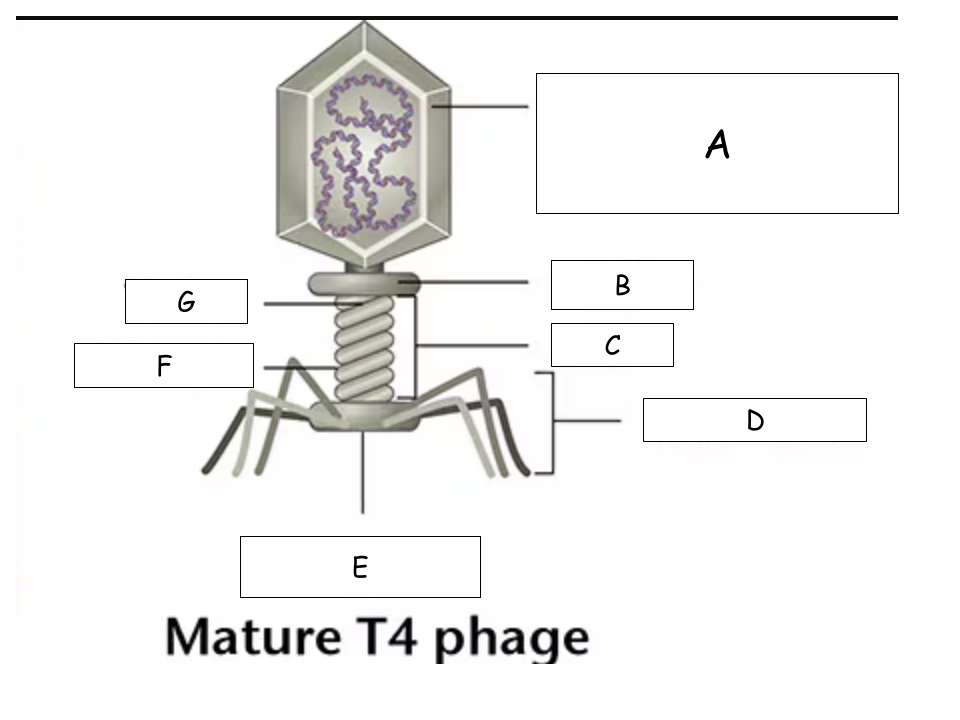
(See Mature T4 Phage.png) F?
Sheath
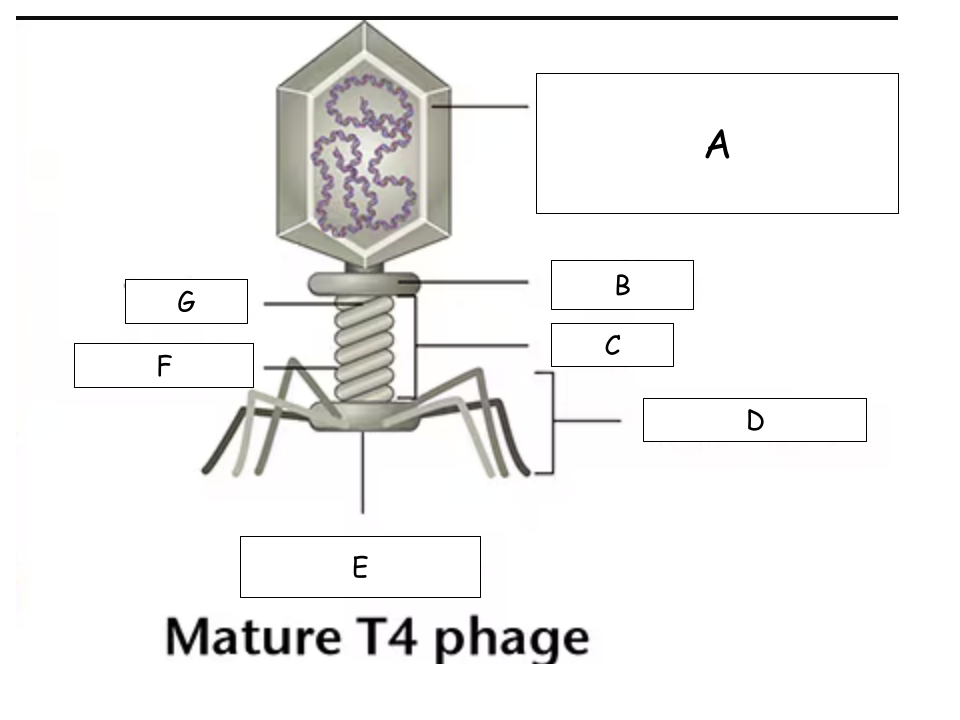
(See Mature T4 Phage.png) G?
Tube
Bacteriophage?
A virus that infects bacteria, using it as the host for reproduction
Transduction?
The process by which foreign DNA is introduced into a cell by a virus or viral vector
Lytic?
Of a bacteriophage, one that infects the cell, replicates, and lyses the cell.
Lysogenic?
Of a bacteriophage, one that infects the cell, but the phage DNA integrates into the host cell genome without replication. (No immediate cell lysis.)
Or, of a bacteria, one that harbors a temperate phage integrated into its chromosome.
Describe the steps of the lytic replication cycle.
Phage adsorbs to the host cell.
Phage DNA is injected, host DNA is degraded.
Phage DNA is replicated, phage parts are synthesized.
Mature phages are assembled.
Host cell is lysed, new phages are released.
Rinse and Repeat
Plaque assay?
A quantitative assay using a serial dilution of a solution containing a large unknown number of bacteriophages that determines the original density (phages per mL)
Plaque?
On an otherwise clear bacterial lawn, a clear area caused by the growth and reproduction of a single bacteriophage.
Formula for initial phage density?
Initial phage density in phages/mL = dilution factor * (plaque number/mL)
Prophage?
The viral DNA that integrates into the bacterial chromosome.
Temperate phages?
Viruses that can become a prophage, integrating DNA into the chromosome of the host bacterial cell and making the later lysogenic.
Virulent phages?
A bacteriophage that infects, replicates within, and lyses bacterial cells, releasing new phages.
Describe the initial finding that led to the Lederburg-Zinder Experiment
The two auxotrophic strains used were called LA-22 and LA-2.
LA-22 could not produce phe and trp,
LA-2 could not produce met and his.
They were initially mixed, producing prototrophs at a rate of 1 prototroph for every 100,000 cells, initially thought to be a result of conjugation.
After replicating the Davis U-tube experiment however, prototrophs still appeared, but only on the side containing LA-22.
Therefore, conjugation couldn’t have been responsible for the creation of prototrophs.
What were the three observations that Lederberg and Zinder saw during their experiment?
The LA-2 cells only produced FA if they were grown in association with the LA-22 cells. Nothing happened if they were grown separately.
Adding DNA-ase waepek sa FA. Therefore, FA is not naked DNA. (→ not transformation)
Changing the filter size (smaller than bacteriophage size) so that FA can’t cross → no recombination
What was the reason behind the recombination in LA-22 cells?
Short answer: P22 bacteriophages.
Long answer:
A bacteriophage called P22 are present initially as a prophage in LA-22 Chromosomes.
After these phages reproduce, lyse the LA-22 chromosome, and are released, they cross the filter and infect and lyse some of the LA-2 cells.
In the process of lysis of LA-2, the P22 will occasionally package a region of the LA-2 chromosome. This region may include phe+ and trp+ genes.
If the P22 phages cross the filter one more time, they could infect the La-22 cells, and lysogenize them. The result are lysogenized cells with the new wild type genes integrated that behave as prototrophs.
How does transduction help us map genes?
Genes that are close enough together can be cotransduced. These genes are thus known to be linked.
Cell stressors?
Conditions that induce viral DNA to leave the chromosome, initiate replication, phage reproduction and lysis.
How are phages helping us solve the problem of ABx-resistant bacteria?
Phages kill bacteria that have adapted to survive antibiotics.
AQ: What type of growth medium contains inorganic salts and a carbon source, but no other organic molecules?
Minimal medium
Heteroduplex?
A double-stranded nucleic acid molecule that is formed when genetic material from different sources recombine.
Ex: One strand is wild type from the host and the other is mutated and foreign. Though homologous, when wound together they form mismatches.
Merozygote?
A bacterial cell that is partially diploid.
AQ: Distinguish between bacterial HGT and VGT
HGT: The movement of genes from one bacterial species to a different species.
VGT: Bacterial genes move from one member of a species to another member of the same species.