RNA to Protein
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
What is translation
converting the mRNA sequence into the language of amino acids
Follows “genetic code” rules
RNA copies of DNA segments direct synthesis of protein
Used by ALL present-day organisms (except the mitochondria)
Requires adaptor molecule (tRNA)
What is an anticodon
a three-nucleotide sequence on a tRNA molecule that is complementary to a specific codon on mRNA
What is a codon
a sequence of three consecutive nucleotides (base pairs) in mRNA that codes for a specific amino acid or signals the termination of protein synthesis
Read in mRNA
First 2 positions of the codon MUST match with the anticodon correctly (3rd can be mismatched)
64 possible combinations of codons, but 20 amino acids
What are eukaryotic initiation factors
Protein that helps load initiator tRNA on to the ribosome, thus initiating translation (eIF)
What is the genetic code
set of rules followed during translation
Ex: AAA = Lys or UGC = Cys
Redundant because of the wobble on the 3rd base
Variations:
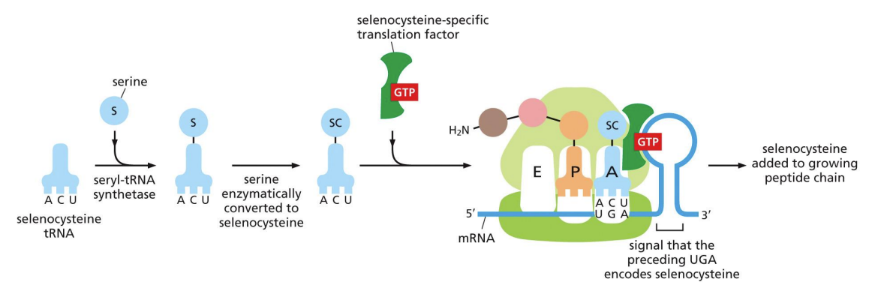
Translation recoding = 21st amino acid can be inserted directly into growing polypeptide using selenocysteine method
Seleoncysteine = produced from serine tRNA that is enzymatically altered
SC tRNA binds to STOP codon, translating the codon into SC
Translational frameshift = similar to leaky scanning (allows more than one protein to be synthesized from a single mRNA by changing the reading frame by ignoring a start or stop codon)
Tends to happen in viruses only
What is wobble
3rd position in a codon that doesn’t need to match with the anticodon
Runs a risk of mutation
Wobble codon base | Possible anticodon bases |
U | A, G, or I |
C | G or I |
A | U |
G | C |
What is an initiator tRNA
the first tRNA that begins translation at AUG codon
All new polypeptide chains start with MET (from initiator tRNA)
AA is eventually removed by protease
What is the reading frame
allow an RNA sequence to be translated in any one of three different frames
Only one of the three is an actual mRNA that encodes for the required protein
Depends on where the frame starts
Start point is crucial to determine whole reading frame
What is the proteasome (simple)
a cylinder that contains proteases (inside) for digesting proteins
What is the ribosome
made of 50 different ribosomal proteins and several rRNAs
Made of 2 subunits
1 large = catalyzes the formation of peptide bonds between amino acids
1 small = important for matching tRNA to codon on mRNA
Matches anticodon to codon
Adds the amino acids to the polypeptide chain
What is a ribozyme
catalytic activity found in RNA molecules
rRNA is responsible for catalytic activity in forming covalent peptide bonds (NOT PROTEINS)
Responsible for catalytic activity by donating an OH to the active site when forming covalent peptide bonds (NOT Proteins)
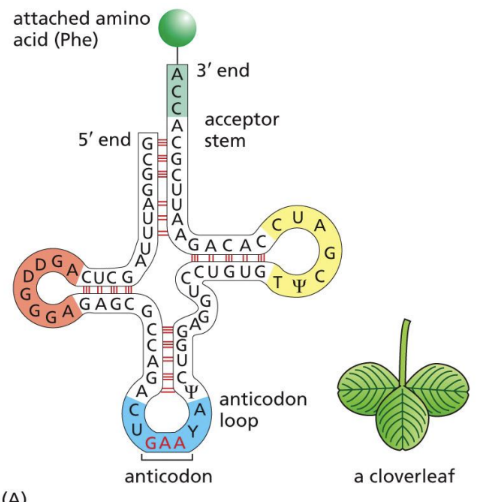
What is a tRNA
Small set of RNA molecules that acts as an adaptor molecule during translation
80 nucleotides long & synthesized by RNA pol. III in nucleus
Splicing process has NO lariat formation (only cut and paste)
~50 different tRNAs because of chemical modifications of bases
Not one per amino
Responsible for catalytic activity by donating an OH to the active site when forming covalent peptide bonds (NOT Proteins)
Each tRNA is unique and is synthesized by a AA-tRNA synthetase
Ensures proper matching of anticodon to amino acid
2 critical regions of unpaired nucleotides
Anticodon = complementary to mRNA codon
Amino acid attachment site at 3’ end
What is ubiquitin
highly conserved eukaryotic protein with many functions (ex: serves as a marker for proteasomes)
What is a nonsense mutation
Premature stop mutation
What is a missense mutation
A different amino acid at a different location
What is a start codon
AUG (Met) bound to tRNA (required to begin translation)
What is a stop codon
UAA, UAG, UGA
What is a ribosome and it’s function
Add the amino acids to the polypeptide chain
Has 4 binding sites for RNA
1 for mRNA
A & P site for tRNAs
Hold tRNA tightly if successfully found a complementary anti-codon to codon
Sites are close together in order to avoid changing the readframe (tRNAs stay next to each other)
E site to exit the tRNA
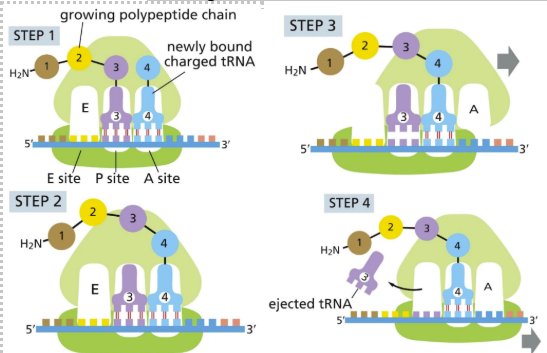
4 major steps to chain elongation:
tRNA binding
Peptide bond formation
Large subunit translocation
Small subunit translocation
What is the process of adding an amino acid to a tRNA
tRNA synthetase:
tRNA requires aminoacyl-tRNA (AA-tRNA) synthetase to recognize and attach the correct amino acid on the 3’ end
20 diff. AA-tRNA synthetases – one for each specific amino acid
Reaction coupled with ATP hydrolysis
Uses ATP to hydrolyze bonds and add the correct amino acid
Must recognize the matching tRNA anticodon
Contains 3 adjacent nucleotide binding pockets (each with complementary shape and charge to anticodon nucleotide)
Several positions on tRNA are important for synthetase recognition (ex: D loop)
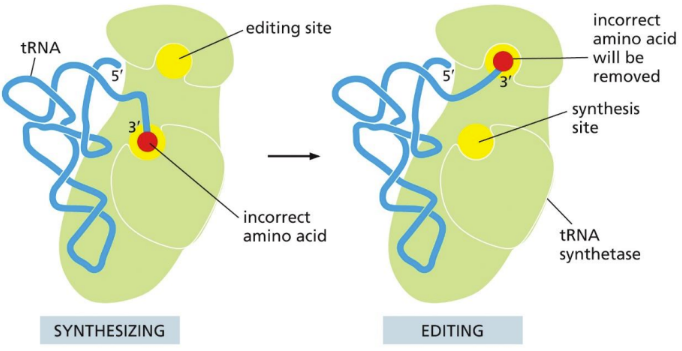
Two editing mechanisms that ensures the correct amino acid to tRNA synthetase
Active site on enzyme has high affinity for the appropriate amino acid
A second proofreading mechanism moves bound amino acid into an editing site (if it fits, it’s rejected by the editing site)
Describe the translation process
Appropriate start sequence, necessary to determine reading frame
With special initiator tRNA at AUG
Initiation factors recognize initiator tRNA, allowing synthesis to begin
Steps
Initiator tRNA (MET) binds to P-site on small subunit ribosome with the help eIF
Small ribosome binds to 5’ end of mRNA (has it’s own eIFs)
Small ribosome moves along mRNA until it finds an AUG
driven by ATP
leaky scanning (helps find correct AUG)
eIF2 leaves small ribosome. Allowing it to bind to the large ribosomal subunit (P with MET and free A site)
AA-tRNA binds to A site and protein synthesis begins (peptide synthesis steps)
Peptide synthesis
Aminoacyl-tRNA molecule binds to vacant A site (if codon and anticodon match)
P-site is bound by another tRNA
AA-tRNA binds to mRNA codon in the A-site
mRNA translated in 5’ to 3’ direction (starting with amine (N) terminus)
Amino acids added at C terminus
Carboxyl end of polypeptide chain is released from P site tRNA and new polypeptide bond is formed
Large subunit translocates relative to the small subunit
Sites are hybrid
E & P on large subunit
P and A on small subunit
Small subunit translocates (RESET RIBOSOME)
Moving the mRNA 3 nucleotides to the A-site
What is the amino acid chain forming
Protein chain made N-terminal to C-terminal end
Peptide bonds formed between the carboxyl group at the end of the growing peptide chain and free amino group on incoming amino acid
Growing carboxyl end still covalently attached to tRNA (bond energy used to attach next AA-tRNA)
What are the steps for translation initiation?
initiator tRNA (MET) binds to P-site on small subunit ribosome with the help eIF
Small ribosome binds to 5’ end of mRNA (has it’s own eIFs)
Small ribosome moves along mRNA until it finds an AUG
driven by ATP
leaky scanning (helps find correct AUG)
eIF2 leaves small ribosome. Allowing it to bind to the large ribosomal subunit (P with MET and free A site)
AA-tRNA binds to A site and protein synthesis begins (peptide synthesis steps)
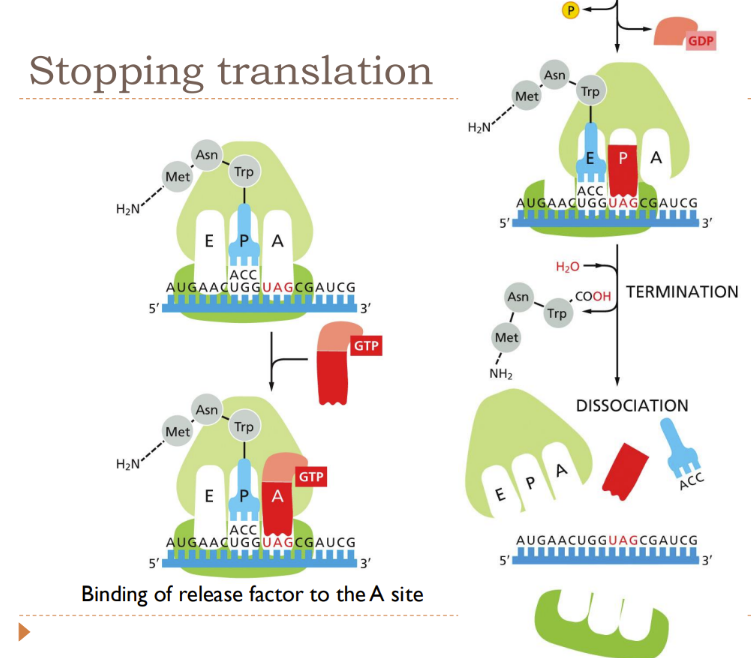
How is translation stopped?
Once a STOP codon is reached (UAA, UAG, or UGA) = they DON’T code for an amino acid
When STOP codon is at the A site, it calls over release factors to enter the rRNA and force the enzymes in the ribosome to add water (NOT an amino acid)
Forms COOH end (C terminus)
The release factor = molecular mimic (similar structure to tRNA => helps release factor bind to A site)
peptide chain is released and begins further folding
Newly transcribed mRNA strand is moved through many ribosomes before degradation
Allows for a lot of proteins to be made from 1 transcript
Protein folding happens immediately as it is being made
What are the steps of peptide chain synthesis?
Aminoacyl-tRNA molecule binds to vacant A site (if codon and anticodon match)
P-site is bound by another tRNA
AA-tRNA binds to mRNA codon in the A-site
mRNA translated in 5’ to 3’ direction (starting with amine (N) terminus)
Amino acids added at C terminus
Carboxyl end of polypeptide chain is released from P site tRNA and new polypeptide bond is formed
Large subunit translocates relative to the small subunit
Sites are hybrid
E & P on large subunit
P and A on small subunit
Small subunit translocates (RESET RIBOSOME)
Moving the mRNA 3 nucleotides to the A-site
What is proofreading
Elongation factors (EF1/2) check tRNA/amino acid pairing for accuracy by providing GTP
If it’s not the correct tRNA, then it will not provide energy (GTP)
& speed up translation process
Helps bring tRNA to the ribosome
Codon-anticodon match checked with GTP hydrolysis (99.99% accuracy)
Small rRNA subunit forms hydrogen bond between the codon and anticodon
If it's a correct match, GTP hydrolysis happens. Moving translation process forward
If it's an incorrect match, tRNA leaves the ribosome
Quality control during translation:
5’ cap and poly A tail = essential for translation initiation
Exon junction complex ensures proper splicing (essential)
Nonsense-mediated mRNA decay (MOST POWERFUL)
When there is more mRNA left to translate but a STOP codon has already been reached (ribosome “senses” there are more EJC therefore, mRNA is destroyed)
Never gets translated into a protein
What are some diseases caused by ribosomopathies
a collection of rare disorders involving impaired ribosome biogenesis and function (mutations affect translation and protein synthesis)
Ex: Diamond Blackfan Anemia, Dyskeratosis Congenita, & Treacher Collins
Alterations in genes encoding ribosomal proteins
Diamond-Blackfan Anemia = congenital mutation in RPS 19 causing hypoplastic macrocytic anemia & bone marrow failure
Alterations in genes involved in ribosomal biogenesis
Myelodysplastic syndrome = congenital mutation similar to DB anemia
Treacher-Collins syndrome = cartilage hair hypoplasia
Ribosomal function abnormalities activate p53 pathway
5q syndrome = entire q arm of chromosome 5 deleted. Presents with severe macrocytic anemia
What are some diseases caused by nonsense mutations
Rare. Caused by truncated protein because STOP codon is misplaced
Some cystic fibrosis mutations
DMD
Beta-thalassemia
Hurler’s syndrome
PTC 124 drug was in clinical trials = makes ribosomes less sensitive to STOP codons
What are some diseases caused by missense mutations
Different amino acid at a different location causes...
Sickle cell disease
Some types of cystic fibrosis mutations
Some cancer types