L2: M/o in medical / pharma assays
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
16 Terms
What are some examples of how m/o are used in medical/ pharmaceutical assays?
Antibiotic assays
Agar diffusion assay, where a filter paper with a specifc conc. of an antibiotic is place on a plate and the organisms susceptibility is tested
Vitamin/amino acid assays uses titration
Absent from medium initially & added in progressively larger concentrations
Greater microbial growth
Removal of antibiotics from pharmaceutical preparations
β-lactamase, produced by certain bacteria & inactivates penicillin (as it targets beta-lactam ring
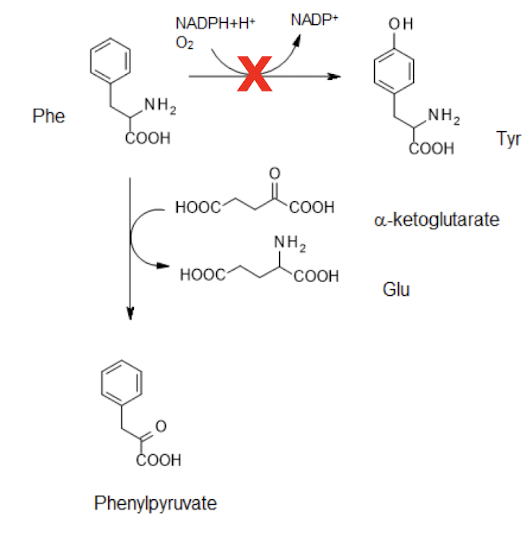
What is PKU (phenylketonuria)?
Inborn error of metabolism where there is an inactive phe hydroxylase
which converts Phe → Tyr
Loss of this results in a transamination between Phe and ⍺-ketoglutarate
Forming PPVA (phenylpyruvate plus Glu)
Controlled by restricting Phe in diet
How are m/o used in PKU testing?
Guthrie test
Heel prick
Bacillus subtilis grown in minimal medium containing inhibitor β-2-
thienylalanine which detects elevated levels of Phe in a newborn's blood sample.
Inhibition alleviated in presence of PPVA or Phe
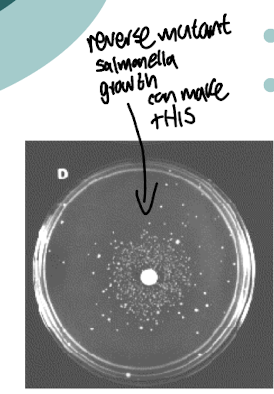
What is the Ames test?
Used to evaluate the mutagenic potential of chemical compounds by observing their effects on non-pathogenic strains of bacteria, often His -strain of Salmonella typhimurium.
Inoculated onto medium containing no His plus a potential mutagen of interest
It measures the rate of mutation to histidine independence in the presence of potential mutagens.
What does growth of S. typhimurium indicate in the Ames test?
Growth of a His- S. typhimurium indicates that there are revertants
Mutated back to wild type (His +), suggesting the compound is a mutagen
Rat liver homogenate can be also incorporated into the test
Determines if a chemical is transformed into a mutagen after transformation in the liver
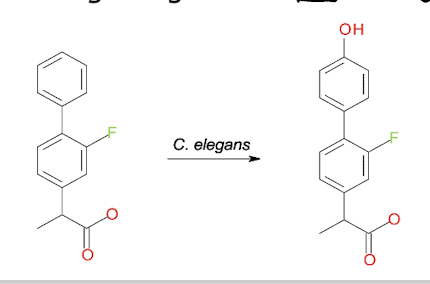
What is an example of using m/o as a model of mammalian drug metabolism?
E.g. Cunninghamella elegans
Fungus that has similar detoxification enzymes to mammals
e.g. Cytochromes P450, conjugative enzymes (add sugar residues to make compounds more water soluble + hydrophilic)
Identical metabolites detected e.g. Flurbiprofen, both mammals and C. elegans generate 4’- hydroxyflurbiprofen as a metabolite which helps in studying the metabolism of pharmaceuticals and their effects on human health.
What are early methods of finding industrially valuable m/o?
Enrichment culture as developed by Beijerinck to isolate specific microorganisms from natural samples by providing selective growth conditions, thereby promoting the growth of desired organisms while inhibiting others.
Serendipitous discoveries e.g. discovery of penicillin in P. notatum by Fleming, which were achieved by chance encounters with useful microorganisms: P. chrysogenum was later found by screening to be 200x more productive
What is the crowded plate technique?
A method used to isolate microorganisms by spreading a diluted sample on a solid growth medium i.e. a soil sample allowing colonies to develop
making it easier to identify and select specific organisms, i.e. by inspecting zones of clearing and isolating single colonies
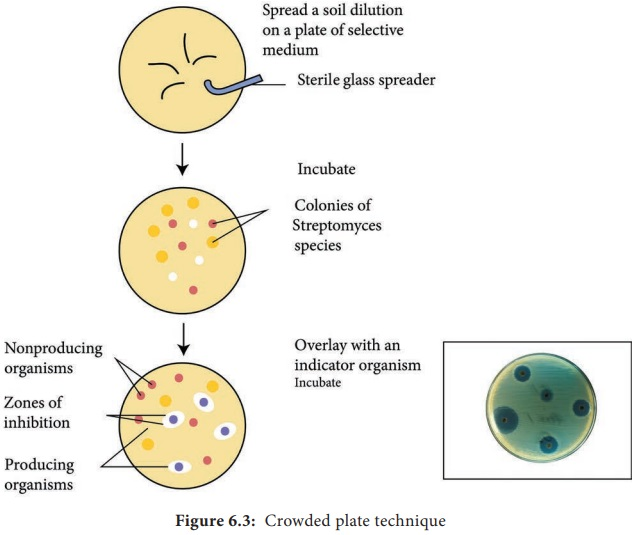
What are advantages and disadvantages of the crowded plate technique?
Advantages: Demonstrably effective, straightforward, relatively quick, inexpensive
Disadvantages:
Limited to easily cultivated strains in known lab/media conditions
High rate of re-discovery due to limited conditions
Un-natural growth conditions, not indicative of the organisms natural environment as they are often overfed with the best combination of nutrients so only the organisms that thrive in these conditions are isolated.
How can we screen for antibiotic producing m/o that are otherwise uncultivable?
Using iChip technology which enables screening of hard to cultivate m/o by using culture conditions that are similar to their environment in situ, allowing researchers to identify antibiotic producers directly from environmental samples.
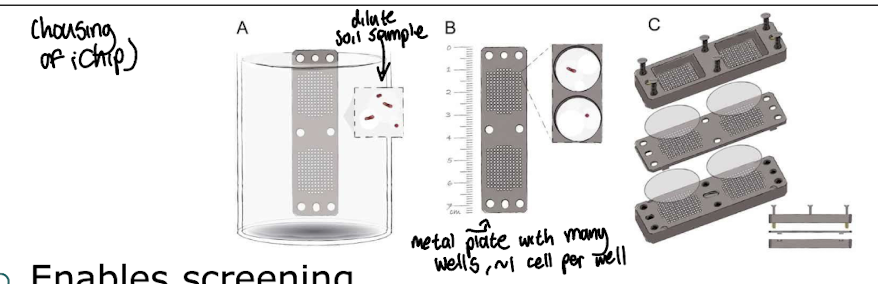
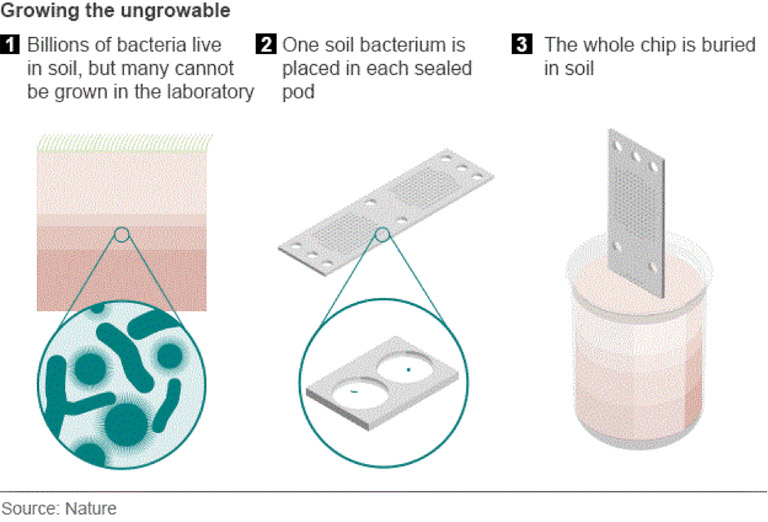
How are the best microbial enzymes for industry discovered?
Traditional screening approaches (isolating microbes from natural sources)
Non-culturing approaches i.e. Metagenomics (looking at all of the DNA sequences in an environment and looking for microbes that produce known genes linked with encoding enzymes of interest
What are traditional methods of screening for enzymes?
Screening on plates coated with starch and stained with iodine i.e. for ⍺-amylase activity, where if a m/o produces the enzyme, there will be a clear zone distinct from the darkened staining around the colony indicating the enzymatic breakdown of starch.
Testing for protease activity on a plate with casein proteins
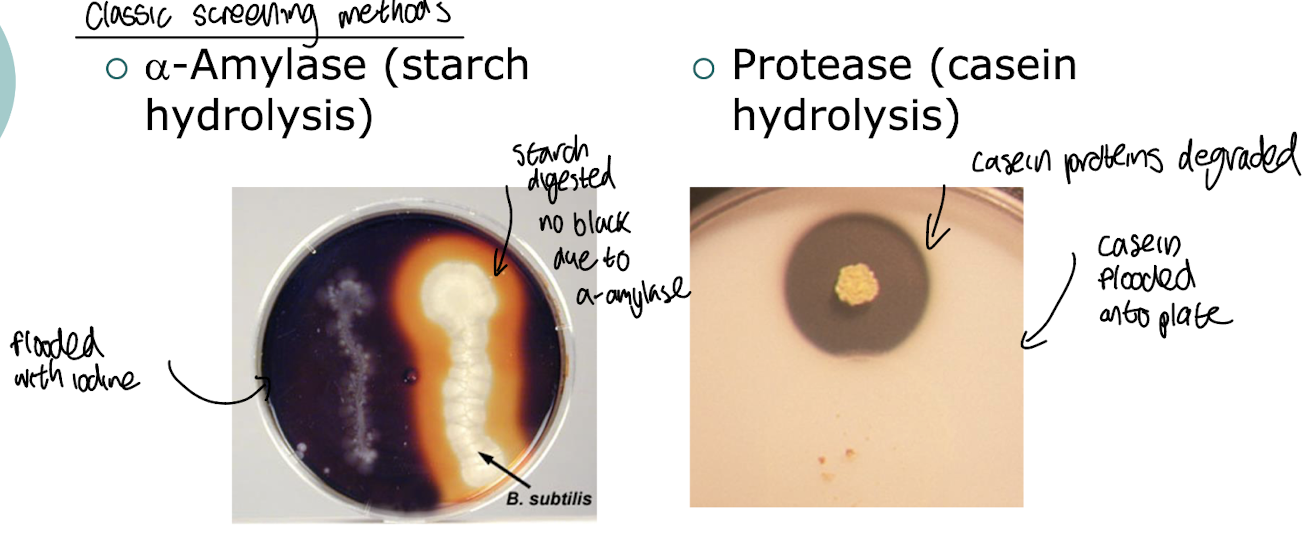
What are features of an effective screen of microbes for a compound of interest?
Easily detectable screening i.e. visual differences on a plate such as colour changes or clear zones indicating metabolic activity
The ability to test multiple samples simultaneously to increase throughput i.e. via Miniaturisation using microtitre plates
Lowered cost and time efficiency, enabling rapid identification of microbial candidates.
How can we summarise the steps of traditional screening?
Choose appropriate environment as source of microbes
Crude primary screen
Solid medium; easily detectable response
Zone of clearing, colour change
Isolate strains of interest
Secondary screening
Cultivate strains in liquid culture
Quantify activity
Isolation of product for characterisation
Optimisation/Scale up if successful in secondary screening, economically, physically viable and stable
How can non-culturable m/o be used?
Access this potential via molecular methods
⚫ Extract DNA directly from environment
⚫ Clone, sequence
⚫ Identify genes through sequence homology
⚫ Metagenomics/bioprospecting
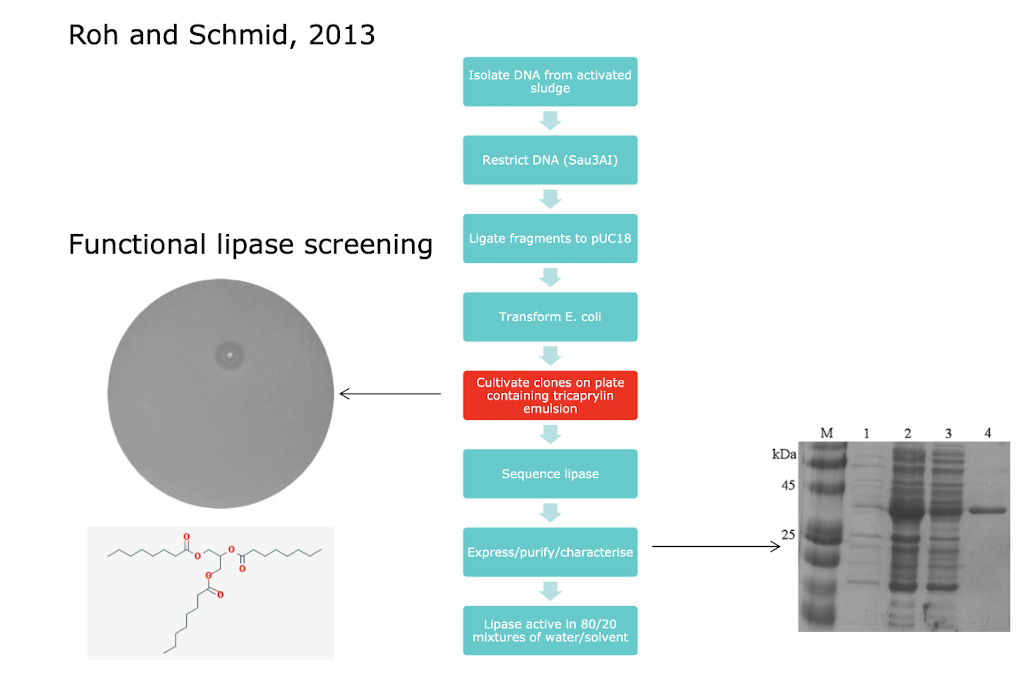
What is an example of a metagenomic approach to isolating a new enzyme?
i.e. Lipase - Reversibly hydrolyse lipids to fatty acid and alcohol
Industrially relevant (fine chemical synthesis; biodiesel)
Barrier to wider use is activity in organic solvents
Typically, chemical reactions are not conducted in aqueous solution (unlike enzymes)
Extensive screening and enzyme engineering efforts to improve solvent tolerance