Chapter 6 : RNA Synthesis and Processing
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
68 Terms
transcription
first step in gene expression
RNA polymerase
enzyme that catalyzes the synthesis of RNA from a DNA template
number of subunits in bacterial RNA polymerase
five
function of the σ subunit in bacterial RNA polymerase
identifies the correct sites for transcription initiation and binds first
promoter
gene sequence upstream of transcription start site to which RNA polymerase initially binds
promoter length in prokaryotes
6 nucleotides
prokaryote promoter locations
10 and 35 base pairs upstream of the transcription start site
bacterial RNA polymerase unwinds this many bases of DNA for transcription
12 - 14
the σ subunit is released after:
addition of ~10 nucleotides
function of the ß and ß’ subunits in bacterial RNA polymerase
form a claw-like structure that grips the DNA template
stop signal
terminates transcription in prokaryotic RNA synthesis
most common stop signal in E. coli
repeat of a GC-rich sequence followed by 7 A residues
result of transcribing the GC-rich repeat in E. coli
RNA forms a stable stem-loop structure
chromatin
location of transcription in eukaryotes
number of RNA polymerase in eukaryotes
3
RNA polymerase II
synthesizes mRNA
RNA polymerase III
synthesizes tRNA and rRNA 5S
RNA polymerase I
synthesizes rRNA 5.8S, rRNA 18S, and rRNA 28S
general transcription factors
proteins involved in transcription from polymerase II promoters
TATA box
promoter for RNA polymerase II that resembles the -10 sequence of bacterial promoters
5 general transcription factors that form the preinitiation complex
TFIID, TFIIB, TFIIF, TFIIE, TFIIH
TFIID
general transcription factor that initially binds the TATA box (or other promotor region)
mediator
protein complex of more than 20 subunits that interacts with both the general transcription factors and RNA polymerase II
process that starts gene transcription
phosphorylation of the polymerase CTD by the TFIIH protein kinase
process that stops gene transcription
RNA polymerase II reaches the terminal signal
process that follows transcription of the terminal signal
RNA endonuclease cleaves RNA polymerase II from the DNA
number of transcription factors that recruit RNA polymerase I
2
general ribosome structure
1 small subunit + 1 large subunit
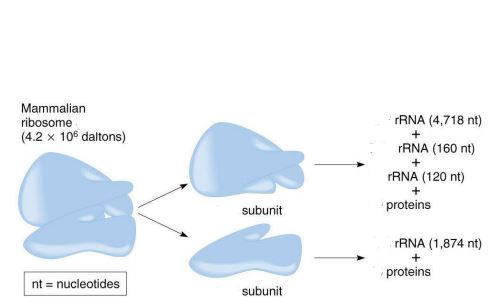
size of the mammalian ribosome
80S
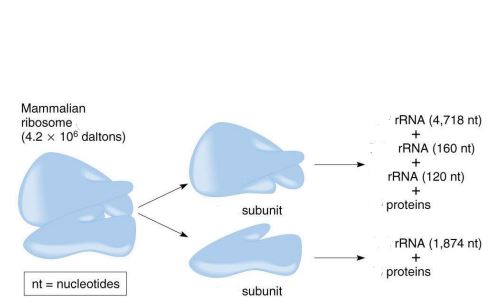
size of the large subunit in a mammalian ribosome
60S
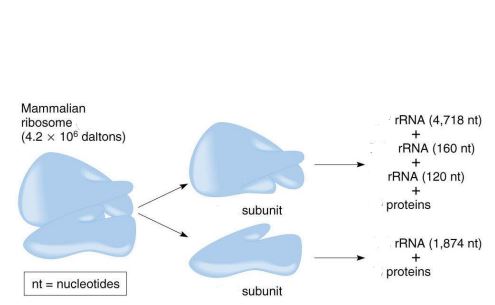
size of the small subunit in a mammalian ribosome
40S
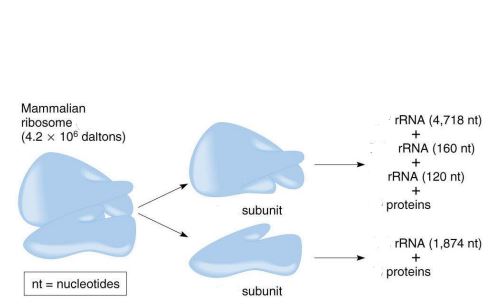
components of the large subunit
28S rRNA + 5.8S rRNA + 5S rRNA + 49 proteins
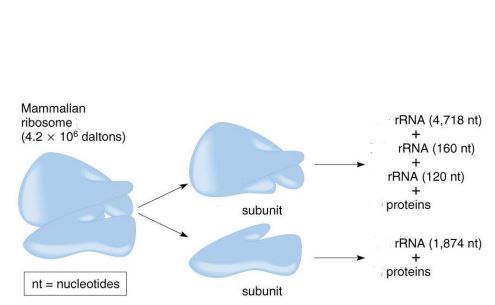
components of the small subunit
18S rRNA + 33 proteins
number of promoter types for polymerase III
3
1st step of pre-rRNA modification
cleavage of rRNA
2nd step of pre-rRNA modification
ribose methylation
3rd step of pre-rRNA modification
uridine isomerization to pseudouridine
snoRNA function
guides modification of pre-rRNA
snoRNP
RNA/protein complex formed with snoRNA to carry out modification
processing of the 5’ end of pre-tRNA
cleavage by the ribozyme RNase P
processing of the 3’ end of tRNA
addition of a CCA terminus
modified bases formed by modification of uridines in tRNA
dihydrouridine and ribothymidine
modified bases formed by modification of guanosines in tRNA
inosine and methylguanosine
when are pre-mRNAs extensively modified
before export from the nucleus
addition of a 7-methylguanosine cap
modification of the 5’ end on mRNA
polyadenylation (addition of a poly-A tail)
modification of the 3’ end on mRNA
G-U rich downstream sequence
signal for polyadenylation
hexanucleotide (AAUAAA)
signal for polyadenylation
number of adenines added by poly-A polymerase to form the poly-A tail
200
1st step of pre-mRNA splicing
cleavage at the 5’ splice site, loop forms by joining the 5’ end of the intron to an A within the intron (branch point)
2nd step of pre-mRNA splicing
cleavage at the 3’ splice site, simultaneous excision of the exons from the loop
spliceosomes
five types of snRNAs
5 types of spliceosomes
U1, U2, U4, U5, U6
snRNP structure
6 - 10 protein molecules complexed with a snRNA
this snRNA binds to the 5’ splice site of pre-mRNA by base pairing recognition
U1
this snRNA binds to the branch point of pre-mRNA
U2
these snRNAs act together to form the intron loop of pre-mRNA
U4, U5, U6
fate of apolipoprotein B when translated from unedited mRNA
synthesized in the liver
fate of apolipoprotein B when translated from edited mRNA
synthesized in the intestine
turnover rate of rRNA
stable (lasts a long time)
turnover rate of tRNA
stable (lasts a long time)
turnover rate of mRNA
very quick; degrades rapidly
short-lived mRNAs code for
regulatory proteins
mRNAs with long half-lives code for
structural proteins
shortening of the poly-A tail
initiates mRNA degradation
regulation of mRNA degradation
extracellular signals, siRNA, miRNA
introns
noncoding sequences
introns are removed at this step of mRNA
splicing