U3 AOS 1 | DNA manipulation
1/116
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
117 Terms
\
channel proteins = molecule cross straight through the membrane.
\
structural word describing a chain
* hormones - insulin to send msgs
* enzymes - speed reactions
* transport - move things
* structure - keratin
* defence - antibodies
\
* there are 20 amino acids
* carboxyl group (COOH)
* NH2 (amine group)
* R - group (varies for each amino acid)
\
polar making them hydrophillic
\
(+) or (-) charged
\
* this creates a polypeptide chain
1. primary - sequence of amino acids which affects overall 3D shape due to the interactions which impacts its function.
\
2. secondary - folding of primary structure into beta pleated sheets, alpha helixes and random coils. alpha helix is flexible and beta pleated sheets provides more structure.
\
3. tertiary - overall 3D shape due to interaction between amino acids and R groups. enables proteins to complete a function
\
4. quarternary - not all go up to this stage. when many polypeptide chains join. creates larger proteins with more structure, complex functions
\
* DNA - deoxyribonucleic acid
* RNA - ribonucleic acid
1. phosphate group
2. pentose (5-carbon) sugar
3. nitrogenous base
\
the pentose sugar is assigned a number in clockwise direction.
* 1’ which attaches to the nitrogenous base
* 3’ which attaches to the phosphate of the following nucleotide
* 5’ which attaches the five-carbon sugar to the phosphate group of the nucleotide.
* pyrimidine: single ring. eg: cytosine, uracil, thymine
* carries the instructions for proteins which are required for cell and organism survival
\
DNA is found in other places besides nucleus; mitochondria and chloroplast have their own DNA.
prokaryotes dont have a nucleus and have a circular chromosome containing DNA.
\
G triple bonds with C (guanine and cytosine)
\
always equal numbers of A and T. and C and G nucleotides
\
DNA polymerase is the enzyme which polymerises DNA
* DNA can be transcribed in one direction (specific)
* enzymes build new strands along template strand by adding nucleotides to 3’ end
\
has uracil which pairs with adenine instead
\
DNA is found in nucleus, mitichondria and chloroplast, RNA found in cytoplasm
\
requires energy
\
* occurs due to the fluid nature of the membrane which allows fusion.
\
1. vesicle contains teh secretory prodycts and is transported to membrane
2. vesicle and plasme membrane fuse
3. the secretory products released from cell into extracellular environment
\
* ribosomes - site of protein synthesis which assemble polypeptide changes by translating mRNA.
* free floating ribosomes make protein for the cell
* ribosomes attached to rough ER make protein for export
* \
* rough ER - if a protein is to be exported, the ribosome creating it is usually attached to the rough ER which allows for folding of the chain before transporting it to the golgi.
\
* transport vesicle - a transport vesicle containing the protein buds off the rough ER and travels to golgi which fuses with golgi membrane and releases it.
\
* Golgi apparatus - proteins can have chemical groups added/removed thus modified where they are packed into secretory vesicles for export or released into cytosol
\
* secretory vesicle - undergoes exocytosis to transport proteins.
\
* plasma membrane which fuses with vesicles and facilitates release of proteins
\
* nucleus which stores DNA which contain the instructions for mRNA.
introns - non coding sections of DNA
exons - coding sections of DNA
termination sequences - signals end of transcription
\
operator region - binding site for repressor proteins
\
leader region - one mode of regulating gene expression
\
^^ in additions to promoter, exons ad termination sequence
1. transcription
\
2. RNA processing
\
3. translation
* occurs in nucleus
\
* DNA unwinds/unzips
* RNA polymerase catalyses transcription through the joining of complementary RNA nucleotides in a 5’ to 3’ end
* transcription of the DNA template strand into pre-mRNA occurs
* pre-mRNA is complementary to the DNA template strand
* in the pre-mRNA, adenine (A) pairs with uracil (U), not with thymine (T).
* in nucleus
\
* addition of methyl G-cap - so bind to ribosome
* adition of poly-A tail - so it cant degrade. adds stability
* introns removed
* exons spliced
* rearranges the order of exons so single gene can give rise to many mRNA strands.
\
* mRNA exits nucleus and binds to ribosome (either in cytosol or rough ER)
\
* ribosome binds to and reads the mRNA molecule
* tRNA anticodons are complementary to the mRNA codons
* tRNA brings the corresponding amino acids to the ribosome
* adjacent amino acids are joined together into a polypeptide chain via a condensation reaction.
\
→ every polypetide chain starts with Met
→ once mRNA reaches STOP codon, no amino acids added
\
* ATP needed
* after translation, polypeptide folds into the protein which can be used in cell or exported out
\
eg: repressor proteins
* conserves energy
* also ensures cells produce approapriate protein
\
* found in prokaryotic cells
* the regulatory gene is always expressed
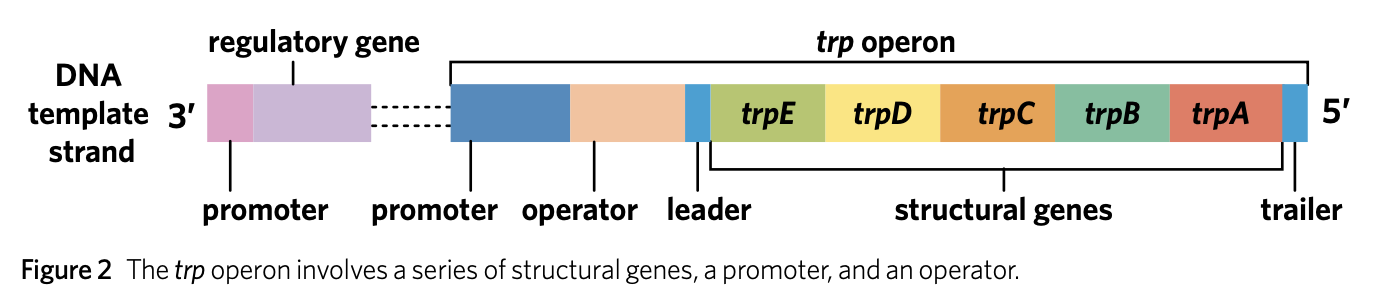
function of the promoter region
The promoter region is the site where RNA polymerase binds to DNA to enable the transcription of the structural genes.
\
* attenuation
repression - high levels of trp
the regulatory gene codes for repressor protein
the tryptophan binds to repressor protein which causes it change shape
the repressor protein is activated and binds to operator which stops RNA polymerase from binding to promoter and transcribing. → inhibits the structural gene
repression - low levels of trp
there is no trp to bind to repressor proteins which makes the repressor protein to becom inactive
this results in RNA polymerase being able to bind to promoter and transcribe the structural genes of trp operon to create enzymes needed for tryptophan production.
* Region 1 has two trp codons.
* The end of the leader region, have attenuator
* Hairpin forms at attenuator
attenuation - high levels of trp
translation and transcription occur simultaneously
ribosme comes up to attenuator sequence
tRNA with trp amino acid travels to ribosome
causes terminator hairpin loop to form
mRNA seperates from template strand and RNA polymerase and ribosome detaches
* ribsome pauses while RNA polymerase continues transcribing as no tRNA has trp to bring to ribosome
* causes mRNA to fold and make antiterminator hairpin loop
* this loop enables RNA polymerase to contine transcribing the structural genes.
→ enzymes are proteins thus made of amino acids
\
* without enzymes, reactions would occur really slowly
* exergonic (releases energy)
* endergonic (absorb energy)
* specific shape - due to interactions between amino acids
* active sit is complementary to substrate
\
induced fit model: active sit changes slghtly to better fit substrate
* active site and substrate bind = enzyme susbtrate complex
* react to form products
activation energy
min amount fo energy needed for reaction to begin
dding more enzymes doesnt lower activation energy further but the reaction DOES speed up as there are more active sites for substrate to bind to.
\
irreversible
optimal temp = fastest rate of reaction
high temp = enzymes denature
* only functions at optimal pH
* saturation point is reached when too many substrates and all active sites are reached = reaction rate os constant after this
* once all substrates are binded, reaction rate stays the same
competitive inhibition
block active site so substrate cant bind
inhibitor has complementary shape to active site
no reaction
increasing substrate can minimise effect of copetitive inhibitors
The more competitive inhibitors = inhibit enzyme activity more = less likely for substrate to bind
\
* they slow rate of reaction but dont stop forever
* can be washed out by dialysis
\
* inhibitor can have its effects overcome by increasing the amount of substrate present
* non competitive, reversible inhibitors arent impacted by substrate conc.
* enzymes will be unable to bind with substrate or catalyse reactions
\
* not proteins but are organic molecules (lipid, carbohydrate).
* reversibly loaded and unloaded
* subset of cofactors
\
during reaction, conenzyme binds to active site and donates energy. substrate also binds.
\
after reaction, coenzymes leave the enzyme and is recycled by accepting more energy to assist other reactions
* each enzyme catalyses a different product which used in the series of reaction
\
* if something goes wrong (eg: enzyme 2 couldn’t function; inhibitor, denaturation, gene mutation) then effects the end product as well as the rest of the reaction)
* ligases
* polymerases
* active site of an endonuclease is complementary to recognition site
* create sticky ends (overhanging nucleotides) or blunt ends (straight thru, no overhangs)
linear DNA - n.o of cuts = fragments (cuts + 1)
\
* RNA polymerase is generally used in transcription
* DNA polymerase is generally used in replication and amplication of DNA
* add to 5’ end
* a primer is required for a polymerase to attach to template strand
* once polymerase attaches to a primer, it reads and synthesises a complementary strand to template in 5’ to 3’ end.
* forensics
* testong for genetic diseases
* taq polymerase (type of DNA polymerase) → used as it has higher optimal temp
* nucleotide bases
* specific DNA primers to join to 3’ end
* restriction enzymes used to cut
1. denaturation - heated to 95 to break H bonds and seperate strands
2. annealing - the strands cooled to 50 so primers can bind
3. extension/elongation - DNA heated to 72 so taq polymerase bonds to primers and synthesises new strand
4. steps 1-3 are repeated to create many DNA strands
gel electrophoresis process
DNA samples loaded into wells using micropipette.
gel is made of agarose - has tiny pres to enable DNA fragments to move
gel is immersed in a buffer to carry the current and maintain temp
electric current passes through the gel using two electrodes
negative electrode is near the wells. - DNA is negatively charged due to its phosphate backbone
when current is applied, the DNA fragments will move from negative to positive
smaller fragments move faster and further than large fragments
large fragments closer to negative electrode
use a UV light or dye to see fragments
* used for comparing sizes
* gel composition
* concentration of buffer
* time
* DNA pofiling, paternity testing
factors that affect migration of DNA fragments through agarose during gel electro
the size of the molecules, as the larger molecule will move more slowly
the charge of the molecule, as the negative charge means that DNA moves towards the positive electrode
the length of time the voltage is applied, as there may not be enough time for the DNA to migrate through the gel
the concentration of the agarose, as denser agarose results in the molecules moving more slowly
\
* replicate independently
\
role of vectors
transport material into another thing
* plasmid vector
* restriction endonuclease
* DNA ligase
1. Plasmid extracted from bacterial cell
2. Same restriction enzyme used to cut the plasmid and gene of interest
3. DNA ligase (creates phosphodiester bonds to join sugar phosphate backbone) used to bind the foreign DNA into plasmid DNA. After binding, the DNA fragment permanent of recombinant plasmid
4. Recombinant plasmid added to bacterial culture via heat shock/electroporation (makes membrane permeable).
5. antibiotic selection to determine which bacteria has been transformed successfully
* Spread antibiotic on petri dish
* The ones with antibiotic resistance gene will survive (recombinant plasmid) .
* The one without the antibiotic resistance will die.
* The ones that die don’t have the recombinant plasmid