Central Dogma and Translation
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
51 Terms
What are the three steps to translational regulation of gene expression
Initiation
Elongation
Termination
Translation is the assembly of WHAT into WHAT
Translation is the assembly of AMINO ACIDS into POLYPEPTIDES
Amino acids contain an WHAT and WHAT group bounded to a central WHAT with a WHAT and a WHAT
Amino acids contain an AMINO and CARBOXYL group bounded to a central CARBON with a HYDROGEN and a R-GROUP
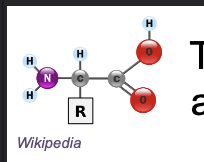
R group is variable and determine WHAT of amino acid
R group is variable and determine UNIQUE CHARACTER of amino acid
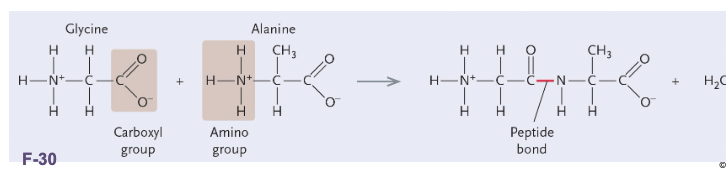
Two amino acids are joined together by a WHAT “WHAT” bond between the WHAT and WHAT by dehydration reaction
Two amino acids are joined together by a COVALENT “PEPTIDE” bond between the AMINO and CARBOXYL by dehydration reaction
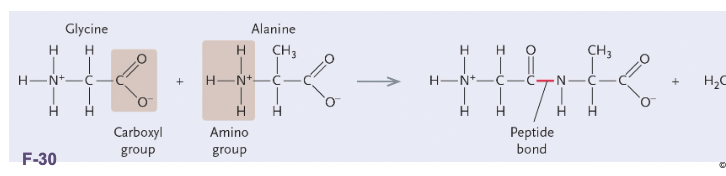
Polypeptides are WHAT chains of amino acids linked by WHAT bonds
Polypeptides are LINEAR chains of amino acids linked by PEPTIDE bonds
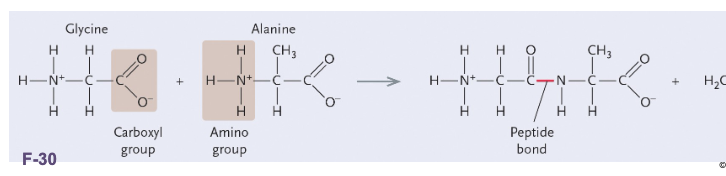
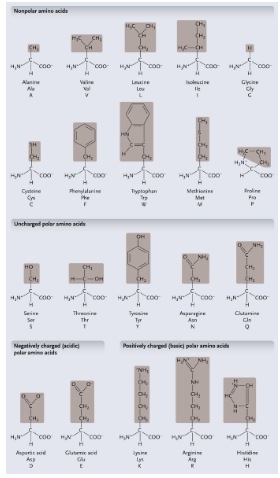
Non-polar amino acids: R groups usually contain WHAT or WHAT
Non-polar amino acids: R groups usually contain CH2 or CH3
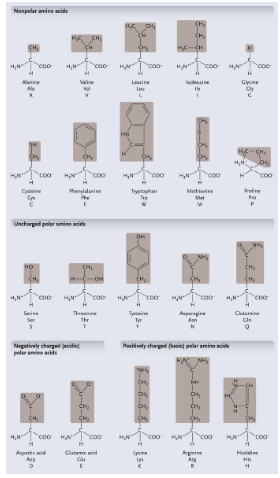
Uncharged polar amino acids: R-group usually contain WHAT (or WHAT)
Uncharged polar amino acids: R-group usually contain OXYGEN (or OH)
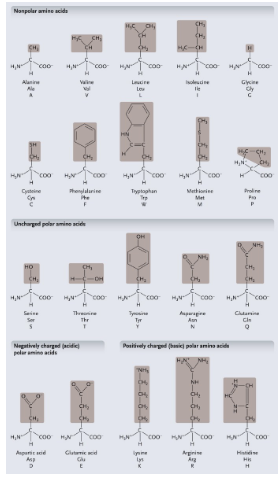
Aromatic amino acids: R group contain a WHAT with alternating WHAT and WHAT bonds
Aromatic amino acids: R group contain a CARBON RING with alternating SINGLE and DOUBLE bonds
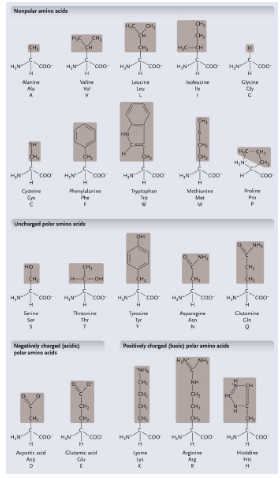
Special functional amino acids include:
WHAT
WHAT
WHAT
Special functional amino acids include:
Methionine
Proline
Cysteine
Methionine: First WHAT added during WHAT
Methionine: First AMINO ACID added during TRANSLATION
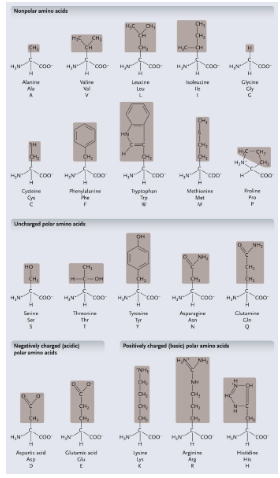
Proline: Causes WHAT (WHAT) in the WHAT
Proline: Causes KINKS (BENDS) in the POLYPEPTIDE
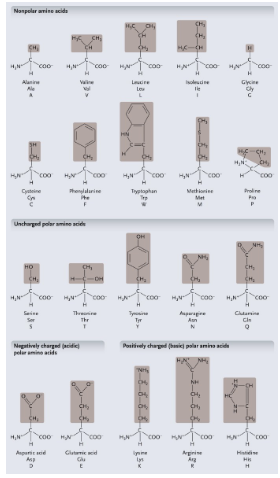
Cysteine: HOW MANY cysteine can form a WHAT between the WHAT atom to make a WHAT
Cysteine: 2 cysteine can form a COVALENT BONDS between the S atom to make a DISULFIDE BRIDGE
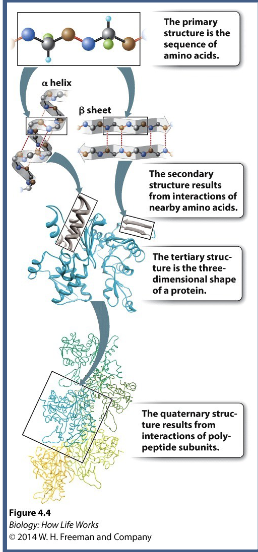
Primary amino acid sequence determines WHAT and WHAT stricture which is critical for proper function
Primary amino acid sequence determines PROTEIN FOLDING and 3-D stricture which is critical for proper function

2nd structure depends on WHAT in the polypeptide WHAT (WHAT and WHAT) sequence WHAT (doesn’t matter about the R-groups or primary structures)
2nd structure depends on HYDROGEN BONDING in the polypeptide BACKBONE (HELICES and SHEETS) sequence INDEPENDENT (doesn’t matter about the R-groups or primary structures)
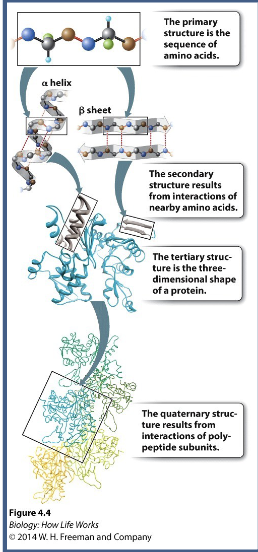
3rd structure is the WHAT structure of a single polypeptide and is composed of interactions between amino acid WHAT → sequence WHAT (does matter about the R-groups)
3rd structure is the 3D structure of a single polypeptide and is composed of interactions between amino acid SIDE CHAINS → sequence DEPENDENT (does matter about the R-groups)
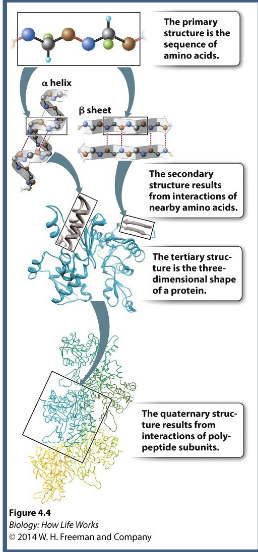
4th structure are interactions between more than one polypeptide to form a WHAT protein (eg, hemoglobin)
4th structure are interactions between more than one polypeptide to form a MULTI-SUBUNIT protein (eg, hemoglobin)

Protein folding disrupted by WHAT (heat and chemicals) or WHAT that change amino acid sequence (how they fold)
Protein folding disrupted by DENATURATION (heat and chemicals) or MUTATIONS that change amino acid sequence (how they fold)
tRNAs: Adaptors between WHAT, WHAT and WHAT
tRNAs: Adaptors between CODONS, TRANSFER RNA and AMINO ACIDS
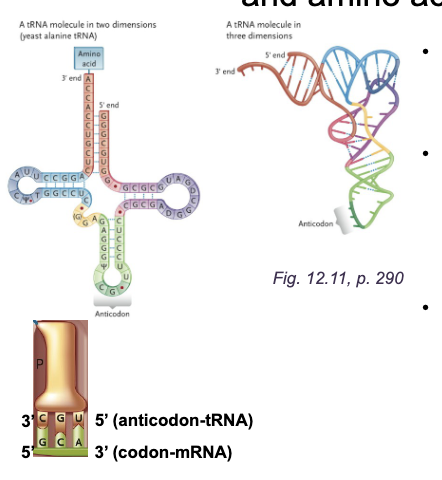
2-D cloverleaf and 3-D L-shaped folded RNA molecule from WHAT
2-D cloverleaf and 3-D L-shaped folded RNA molecule from SELF-COMPLEMENTARITY
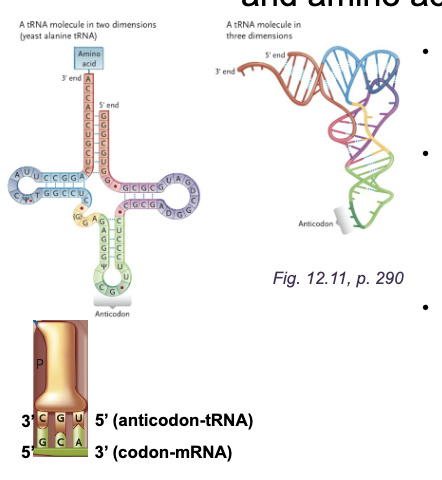
WHAT is where the amino acid is attached and contains the sequence HWAT at the WHAT end of tRNA
ACCEPTOR STEM is where the amino acid is attached and contains the sequence 5’-CCA-3’ at the 3’ end of tRNA
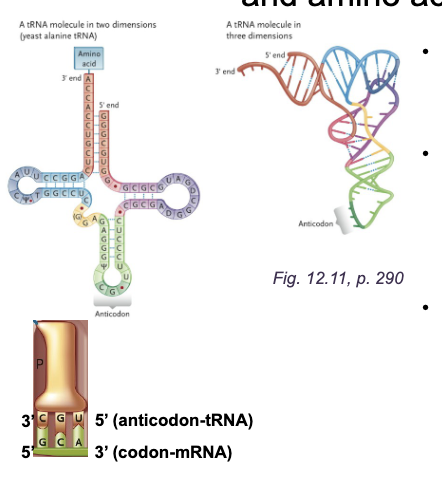
The WHAT is the bottom loop of the cloverleaf and is a WHAT sequence that base pairs to the WHAT on the mRNA
The ANTICODON is the bottom loop of the cloverleaf and is a THREE-NUCLEOTIDE sequence that base pairs to the CODON on the mRNA
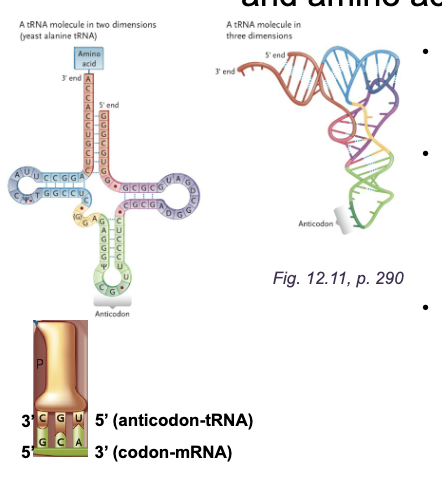
Aminoacyl-tRNA charging
Aminoacyl-tRNA synthetase adds an WHAT to the WHAT of the correct tRNA
There is a unique WHAT for each of the 20 amino acids
Aminoacyl-tRNA synthetase adds an AMINO ACIDS to the ACCEPTOR STEM of the correct tRNA
There is a unique AMINOACYL-tRNA SYNTHETASE for each of the 20 amino acids
Charging reaction (WHAT): WHAT
Charging reaction (AMINOACYLATION):
Amino acid + tRNA + ATP → aminoacyl-tRNA + AMP + PPi
Charging means WHAT
Charging means ADDING an AMINO ACID
Aminoacyl-tRNA charging DIAGRAM
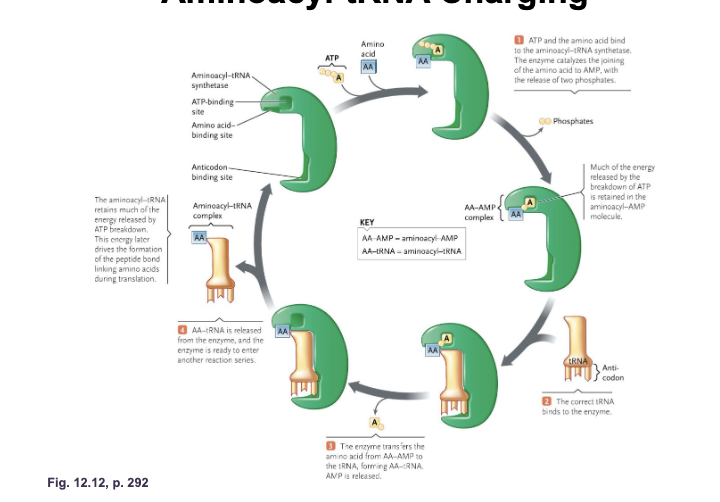
The genetic code consists of WHAT and the WHAT specified by these codons
The genetic code consists of CODONS and the AMINO ACIDS specified by these codons
The codons are written WHAT to WHAT, as they appear in the mRNS:
61 “WHAT” WHAT (code for an amino acid)
3 WHAT/WHAT/WHAT and do NOT code for amino acids (transfer RNA does NOT bind to these codons)
The codons are written 5’ to 3’, as they appear in the mRNS:
61 “SENSE” CODONS (code for an amino acid)
3 TERMINATOR/STOP/NONSENSE and do NOT code for amino acids (transfer RNA does NOT bind to these codons)
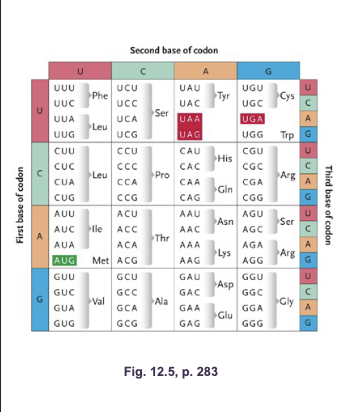
Genetic code shows WHAT in which amino acid can be specified by more than one codon (multiple codes (codons) can make the same amino acid)
Genetic code shows DEGENERACY in which amino acid can be specified by more than one codon (multiple codes (codons) can make the same amino acid)
Codons on the mRNA are read in the WHAT to WHAT direction
Codons on the mRNA are read in the 5’ to 3’ direction
Codons are WHAT and the message contain NO WHAT
Codons are NON-OVERLAPPING and the message contain NO GAPS
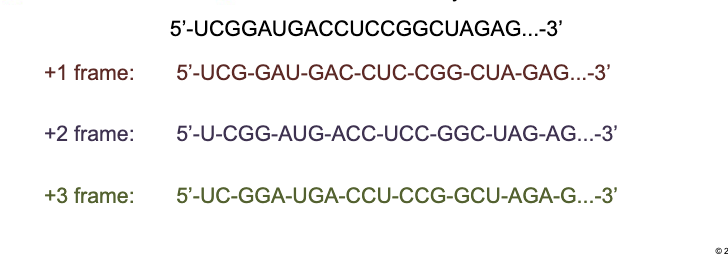
Message is translated in a fixed WHAT set by the WHAT
Message is translated in a fixed READING FRAME set by the START CODON

Amino acids added WHAT to WHAT so the “N” is on the WHAT side and “C” is on the WHAT side
Amino acids added AMINO to CARBOXY so the “N” is on the LEFT side and “C” is on the RIGHT side
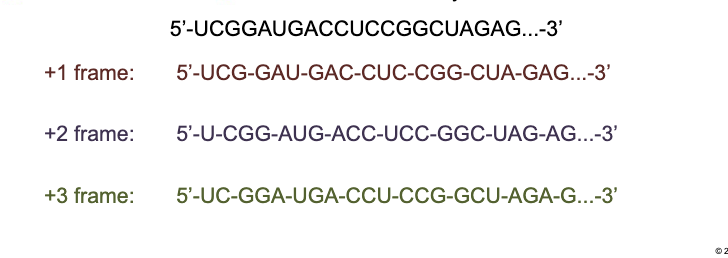
How many reading frames are there in translation
3 reading frames
+1 frame: Splits the code into the codons
+2 frame: Ignores the first letter (first codon)
+3 frame: Ignores the first 2 letters (first two codons)

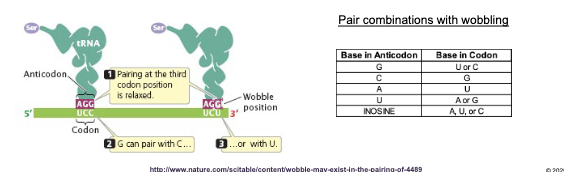
Are there 61 different tRNAs that bond to the 61 mRNA codons? If not why?
No, there are less because some tRNAs read more than one codon

The base at the WHAT end of the WHAT can H-bond with more than one type of base at the WHAT end of a codon: known as WHAT
Pairing of the other two nucleotides in the anticodon with the codon is WHAT
The base at the 5’ end of the ANTICODON can H-bond with more than one type of base at the 3’ end of a codon: known as WOBBLE
Pairing of the other two nucleotides in the anticodon with the codon is PRECISE
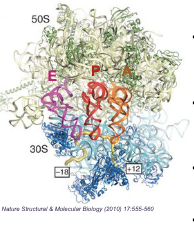
Ribosome protein synthesis machinery
Composed of WHAT made of a complex of WHAT + WHAT
Ribosome protein synthesis machinery
Composed of 2 SUBUNITS made of a complex of rRNA + PROTEINS
Ribosome protein synthesis machinery: What are the 2 subunits
Larger subunit
Small subunit
Ribosome protein synthesis machinery: Large subunit
(WHAT in prokaryotes; WHAT in eukaryotes) Made of WHAT + WHAT and contains the WHAT for formation of peptide bonds
Ribosome protein synthesis machinery: Large subunit
(50S in prokaryotes; 60S in eukaryotes) Made of rRNA + PROTEINS and contains the PEPTIDYL-TRANSFERASE CENTER for formation of peptide bonds
Ribosome protein synthesis machinery: Small subunit
(WHAT in prokaryotes; WHAT in eukaryotes) Made of WHAT + WHAT and contain WHAT where charged WHAT read the codon of mRNA
Ribosome protein synthesis machinery: Small subunit
(30S in prokaryotes; 40S in eukaryotes) Made of rRNA + PROTEIN and contain DECODING CENTER where charged tRNAs read the codon of mRNA

Ribosome protein synthesis machinery
Each subunit exists separately in the WHAT, but join on the WHAT molecule
Ribosome protein synthesis machinery
Each subunit exists separately in the CYTOPLASM, but join on the mRNA molecule

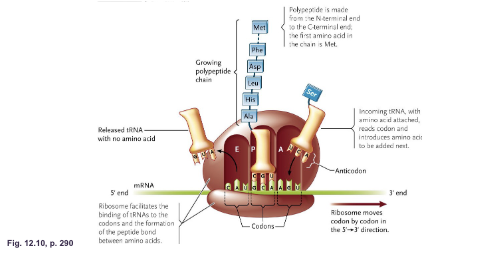
What are the three tRNA binding sites of ribosomes:
WHAT
WHAT
WHAT
What are the three tRNA binding sites of ribosomes:
A site (aminoacyl) (acceptor)
P site (peptidyl)
E site (exit)
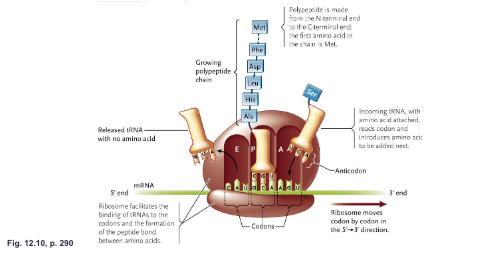
A site (aminoacyl): Binds to the WHAT carrying the WHAT
A site (aminoacyl): Binds to the tRNA carrying the NEXT AMINO ACID

P site (peptidyl): Binds to the WHAT that is attached to the WHAT
P site (peptidyl): Binds to the tRNA that is attached to the GROWING POLYPEPTIDES
E site (exit): Binds to the WHAT that carried the WHAT
E site (exit): Binds to the tRNA that carried the PREVIOUS AMINO ACID
Translation initiation (eukaryotes):
Initiator WHAT and WHAT are brought to the WHAT-site of the WHAT ribosomal subunit
Complex of initiator tRNA(met) and small ribosomal subunit is recruited to the WHAT end of the mRNA and WHAT along the mRNA in a WHAT-WHAT direction until it reaches the WHAT
Complementary base pairing occurs between WHAT of the initiator tRNA(met) and the WHAT codon on the mRNA
Large ribosomal subunit binds small subunit to form the WHAT, which is now ready to accept the next WHAT in the WHAT site
WHAT is hydrolyzed and WHAT begins
Translation initiation (eukaryotes):
Initiator tRNA(met) and GTP (energy) are brought to the P-site of the SMALL ribosomal subunit
Complex of initiator tRNA(met) and small ribosomal subunit is recruited to the CAPPED 5’ end of the mRNA and SCANS along the mRNA in a 5’-3’ direction until it reaches the START CODON (5’ AUG 3’ (mRNA))
Complementary base pairing occurs between ANTICODON of the initiator tRNA(met) and the START codon on the mRNA
Large ribosomal subunit binds small subunit to form the INITIATION COMPLEX, which is now ready to accept the next tRNA in the A site
GTP is hydrolyzed and TRANSLATION begins

Translation: Elongation:
WHAT adds a CHARGED aminoacyl tRNA to the WHAT-site with the hydrolysis of WHAT
The WHAT of the incoming tRNA must be complementary of the WHAT on the mRNA
Translation: Elongation:
ELONGATION FACTOR (EF-GTP) adds a CHARGED aminoacyl tRNA to the A-site with the hydrolysis of GTP
The ANTICODON of the incoming tRNA must be complementary of the CODON on the mRNA
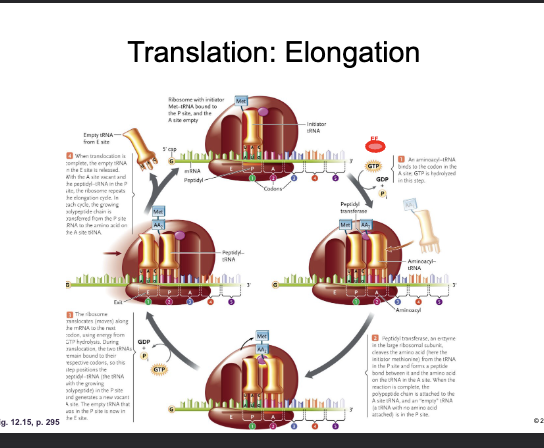
Translation: Elongation:
The WHAT of the large subunit forms a WHAT bond between the WHAT of the amino acid in the WHAT-site and the WHAT of the amino acid in the WHAT-site
The tRNA in P-site is now WHAT (empty)
Translation: Elongation:
The PEPTIDYL TRANSFERASE CENTER of the large subunit forms a PEPTIDE bond between the CARBOXYL of the amino acid in the P-site and the AMINO GROUP of the amino acid in the A-site
The tRNA in P-site is now UNCHARGED (empty)
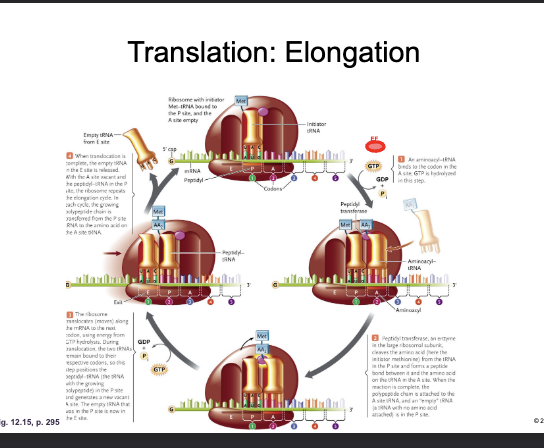
Translation: Elongation
WHAT (shifts) the ribosome three nucleotides along the mRNA (WHAT-WHAT direction)
The empty tRNA is now in the WHAT-site and is WHAT
The tRNA with the growing WHAT is now in the WHAT-site
The WHAT-site is now WHAT (ready to repeat)
Translation: Elongation
GTP HYDROLYSIS TRANSLOCATES (shifts) the ribosome three nucleotides along the mRNA (5’-3’ direction)
The empty tRNA is now in the E-site and is EJECTED
The tRNA with the growing POLYPEPTIDE is now in the P-site
The A-site is now VACANT (ready to repeat)
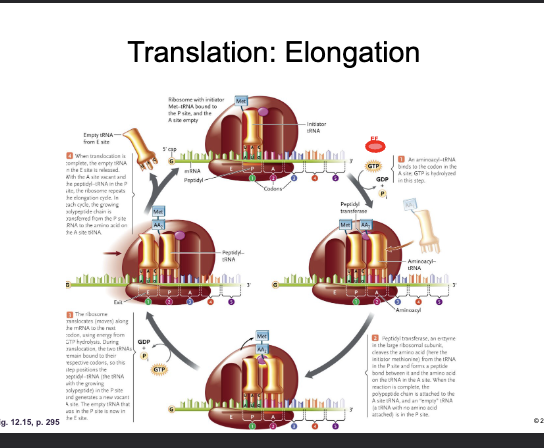
Translation: Elongation
The next codon of the mRNA is now aligned with the WHAT-site and the next WHAT can be loaded by WHAT
Translation: Elongation
The next codon of the mRNA is now aligned with the A-site and the next AMINOACYL tRNA can be loaded by EF-GTP
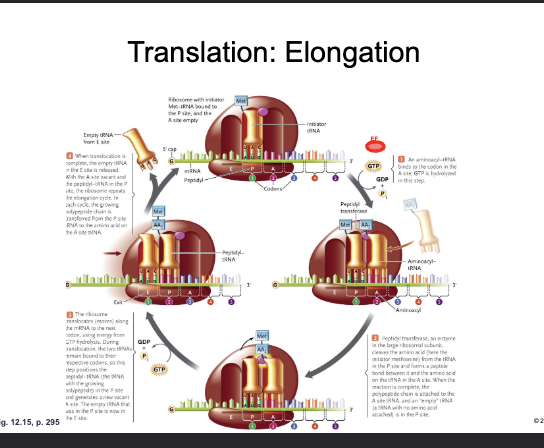
Translation: Termination
One of three WHAT codons are found at the WHAT end of the mRNA: UAA, UAG, UGA
There are NO WHAT for the stop codons
When the ribosome reached the stop codon, WHAT enters the WHAT-site
WHAT stimulates peptidyl transferase to WGAR the polypeptide from the WHAT-site tRNA → the polypeptide is released form the WHAT
The translational machinery WHAT from the mRNA
Translation: Termination
One of three TERMINATION (STOP) codons are found at the 3’ end of the mRNA: UAA, UAG, UGA
There are NO tRNAs for the stop codons
When the ribosome reached the stop codon, RELEASE FACTOR (RF) enters the A-site
RF stimulates peptidyl transferase to CLEAVE the polypeptide from the P-site tRNA → the polypeptide is released form the RIBOSOME
The translational machinery DISSOCIATE from the mRNA
