BASIC GENETICS IN RELATION TO DNA, GENE OGRANISATION, GENE EXPRESSION & POLYMORPHISM
1/68
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
69 Terms
what type of medicine is going to be used increasingly in the future
personalised medicine
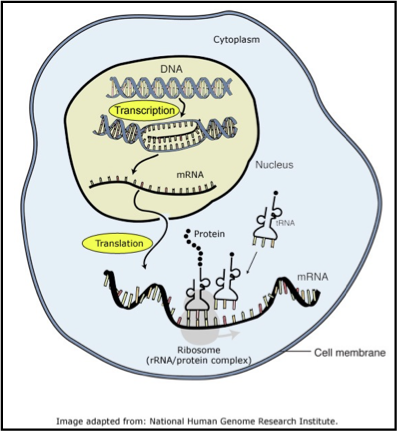
define transcription and translation
transcription: process of copying a segment of DNA into RNA
translation: process of synthesising new protein from messenger RNA
is the genome static/ fixed or dynamic
the genome is static\ fixed
most cells have a copy of the same genome
are cells static/ fixed or dynamic
a cell is dynamic
because they respond to external conditions
most cells follow a cell cycle of division
when do cells differentiate
during development
define pseudogenes
pseudogenes: a DNA sequence/ section of chromosome that is an imperfect copy of a functional gene but is actually non-functional
what percentage of the genome encodes for proteins
3% (21,000 genes)
outline non-coding DNA (‘junk’ DNA)
landing spots for proteins that influence gene activity i.e. switch on or off
strands of RNA with myriad roles
places where chemical modifications silence stretches of chromosome
outline mRNA
shorter than DNA
only contains information coding for one protein or part of a protein
has uracil instead of thymine
single stranded
has a ribose sugar
why does RNA contain uracil instead of thymine
uracil costs a lot less energy to make than thymine
we make a lot of RNA, hence it contains U instead of T
outline transcription
genetic information is only carried on the coding strand of DNA
the other strand is the template strand
strands of DNA that serves as a coding template for one gene may be non-coding for other genes in the same chromosome
the coding strand is always the top strand of DNA
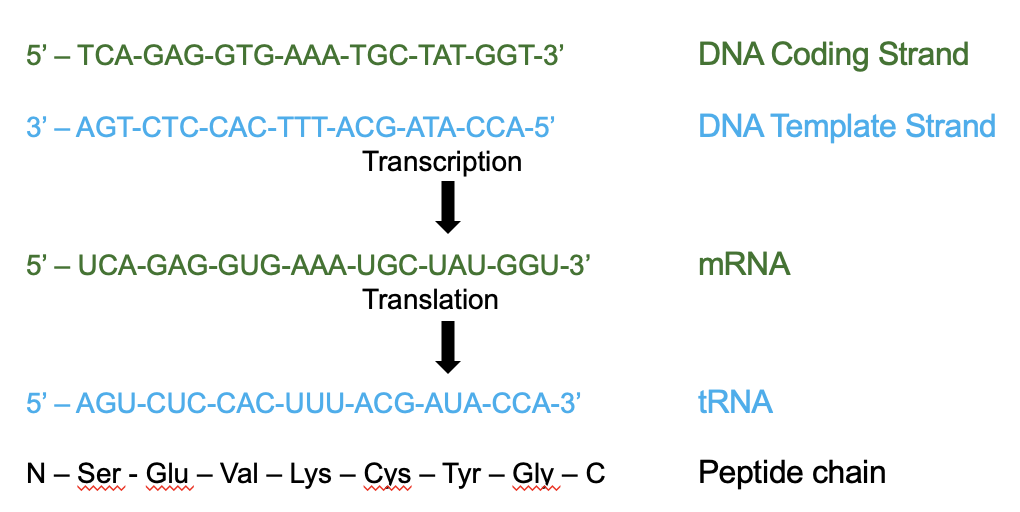
what does this image show
the mRNA sequence is complementary to the DNA template strand and identical to the coding strand (apart from T-U)
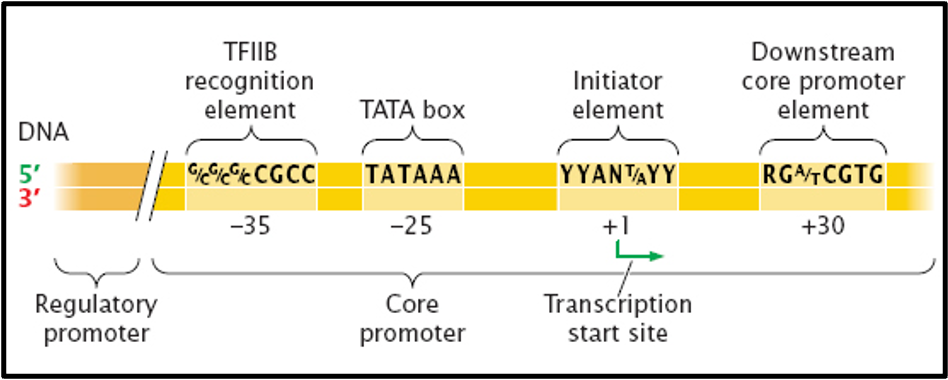
outline initiation
initial step: find the start of the gene on the coding strand of DNA
RNA polymerase must pick the correct strand and find the start of the gene
RNA polymerase binds to one or more short sequences upstream of the start of the gene
closer to the 5’ end
promoter sequences
what are the types of RNA polymerases found in eukaryotes
polymerase I - transcribes large ribosomal RNA
nucleolar region of nucleus
polymerase II - produces mRNA precursors
polymerase III - small RNAs (tRNA), 5S ribosomal RNA and other small DNA sequences
outline RNA polymerase II
produces mRNA precursors
composed of several subunits but requires several accessory proteins (transcription factors)
all added to the complex in a defined order to initiate transcription
RNA polymerase wraps around both strands
large enough to enclose the promoter and beginning of gene
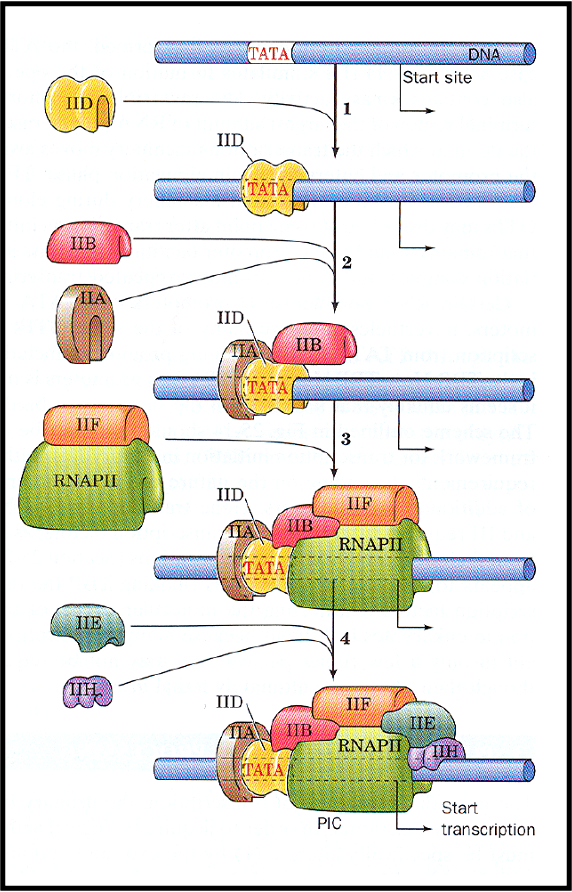
what is TFIID
TFIIID recognises specific region called TATA box
ensures correct placement of RNA polymerase
what is the function of the TATA box
affects transcription rate
determines location of start site
where is the TATA box located
the basal promoter contains the TATA box and is found in all protein-coding genes
define enhancers
enhancers: DNA sequences which control the efficiency and rate of transcription
regulates expression of genes in specific cell types and controls timing of gene expression
why are promoters and enhancers ‘cis acting elements’
they are found on the same molecule (strand) of DNA as the gene they regulate
what characteristic of DNA allows for DNA looping
flexibility of DNA allows for DNA looping
where are promoters located
promoter necessary to start transcription
usually on 5’ side of gene to be transcribed
where are enhancers located
enhancers can affect transcription from afar
on the 5’ or 3’ of transcription start site
on introns or even on non-coding strand
enhancers can be thousands of nucleotides away from interacting promoters
how are enhancers and promoters brought closer together
by the looping of DNA
this occurs due to interactions between proteins bound to enhancers and promoters
what is the name for the proteins facilitating DNA looping
activators
what is the name for proteins that inhibit DNA looping
repressors
what type of promoters and enhancers are there
‘strong’ or ‘weak’ promoters and enhancers
what can changes in promoter strength result in
disease
can also have deleterious effects on a cell
—
e.g. tumour promoting viruses transform healthy cells by inserting strong promoters in vicinity of growth-stimulating genes
e.g. translocation in some cancer cells place genes that should be ‘turned off’ in proximity of strong promoters and enhancers
what do transcription factors bind to
TF bind to promoter and enhancer sequences and recruit RNA polymerase
outline basal transcription factors
required at every promoter site for RNA polymerase interaction - TFIID
why are transcription factors ‘trans’ acting factors
because they are proteins encoded by a different gene to that being regulated
outline transcription after RNA polymerase is attached
RNA polymerase unwinds the double helix over a short length and splits them apart
this creates a bubble of 10 bases
RNA polymerase catalyses sugar phosphate bond between 3’ -OH of ribose and and 5’ PO4
nucleotides are added to the 3’ -OH of growing chain
DNA zips back up as polymerase moves along
the bubble moves along the chain
the growing RNA chain detaches from template
how does the structure of RNA allow it to fold into a 3D structure
RNA is single stranded but base pair interactions between complementary sequences found elsewhere on the same molecule allows 3D folding
folding similarly to the way a polypeptide chain folds into a protein
what does the ability of RNA to fold into complex 3D shapes allow for
allows for some RNA molecules to have structural and catalytic functions (active sites can be formed by RNA folding)
outline pre-mRNA processing
as RNA polymerase moves down the gene
cap on 5’ end - this stabilises the mRNA which is essential for transporting RNA out of the nucleus
alternative splicing
polyA tail on 3’ replaces 3’ UTR causes cleavage at stop codon (AAUAAA)
what is the stop codon needed for
AAUAAA needed for release of polymerase from DNA template
how long is pre-mRNA
pre-mRNA is only 20-40 nucleotides long
when does capping pre-mRNA take place
takes place during transition from transcription initiation to elongation
why does capping pre-mRNA take place
protects from degradation
serves as assembly point for proteins needed to recruit small subunits of ribosomes to begin translation
outline alternative splicing
several variants of the same protein are produced by one gene
mutations at splice sites can result in aberrant or truncated proteins that do not function properly
this plasticity allows for disease development e.g. cancer
what is a spliceosome
spliceosome: exons defined by short, degenerate classical splice site sequences at intron/ exon borders
what are the types of spliceosome
major spliceosome: removes 99.5% of introns
minor spliceosome: removes remaining 0.5% of introns
what are the two main functions of spliceosomes
recognition of intron/ exon boundaries
catalysis of reactions which remove non-coding introns and stitch flanking exons back together
how many different proteins are associated with human spliceosomes
> 300 different proteins
what are mutations that can alter splice sites or spliceosome proteins and what do they result in
mis-splicing » rapid degeneration of mRNA
mis-regulation of splicing factor levels » cancer
list some types of RNA
mRNA
rRNA
tRNA
ncRNA
small nuclear RNA (snRNA)
small nucleolar RNA (snoRNA)
microRNA (miRNA)
outline mRNA
wide range of sizes reflecting size of polypeptide
many common to most cells
‘housekeeping’ proteins needed by all cells e.g. enzymes of glycolysis
specific for only certain types of cells
proteins needed by ‘specialised’ cells e.g. haemoglobin in precursors of RBCs
outline rRNA
builds ribosomes
4 kinds in eukaryotes
what are the 4 kinds of rRNA in eukaryotes
18S - one of these plus other proteins make small subunit
28S, 5.8S, 5S - one each of these plus proteins make a large subunit
S = Svedburg unit (sedimentation rate related to mass and shape)
basically how dense each RNA is
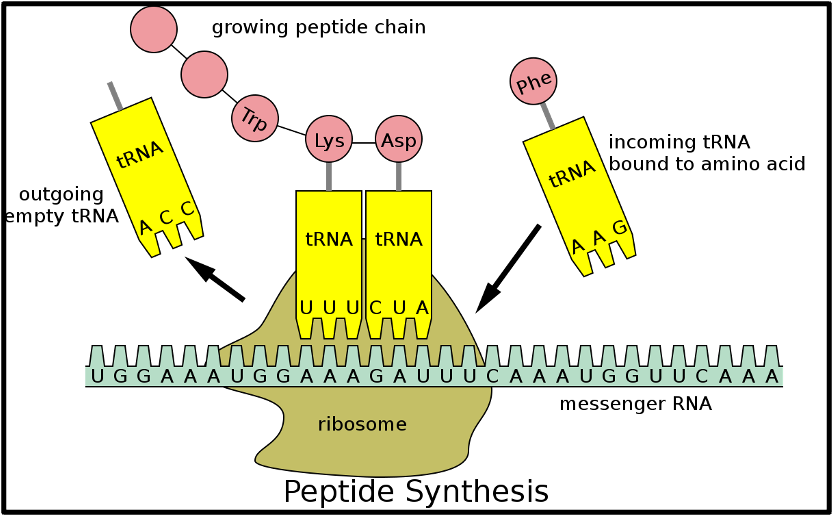
outline tRNA
32 different kinds in typical eukaryotic cells
each kind carries one of 20 amino acids at 3’ end (most amino acids have more than one tRNA)
outline ncRNA
97% of DNA is referred to as junk DNA
90% of genome is actively transcribed and thus more complex
ncRNA can modify protein levels by a mechanism independent of transcription
play major roles in cellular physiology, development, metabolism, implicated in disease process
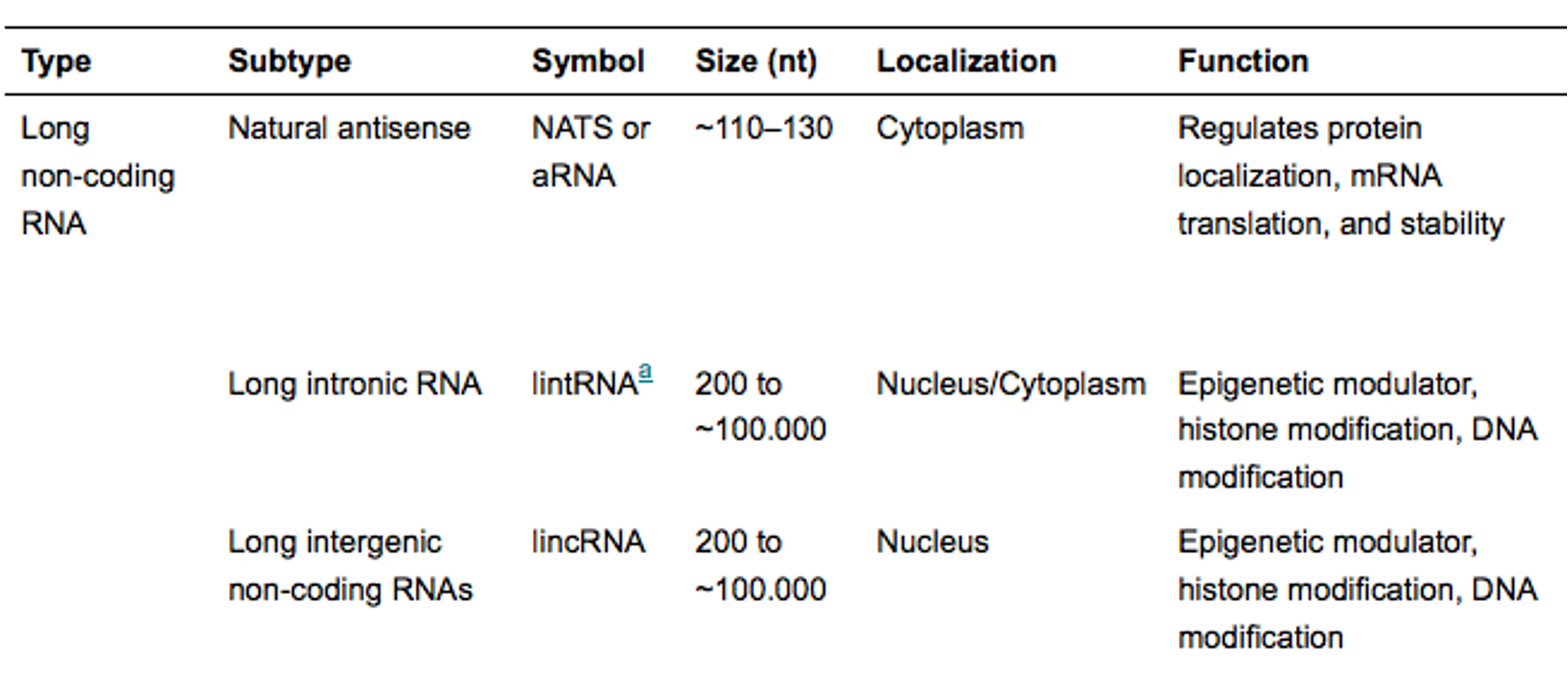
outline long non-coding RNA
long intronic RNA are from cleaved introns from mRNA
outline short non-coding RNA
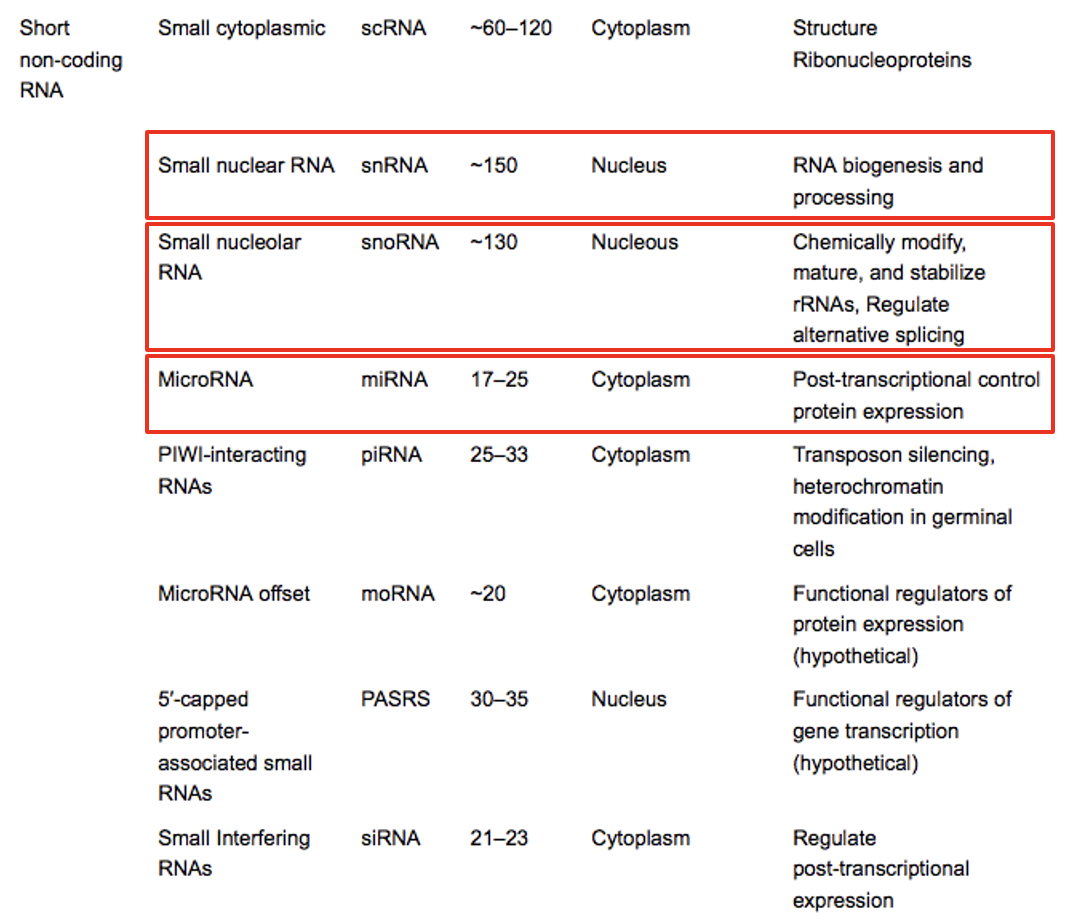
outline small nuclear RNA (snRNA)
several members part of spliceosome
outline small nucleolar RNA (snoRNA)
participate in making ribosomes
cleave precursors of 28S, 18S, 5.8S
implicated in alternative splicing
template for synthesis of telomeres
vertebrates: snoRNA made from cleaved introns
what are telomeres
telomeres: found at the end of chromosomes, areas of long RNA repeat regions thought to protect chromosomes from degradation
gets shorter as we get older
outline microRNA (miRNA)
tiny (18-25 nucleotides)
regulate gene function post-transcriptionally
binds to mRNA and causes degradation
inhibits protein synthesis
regulation of developmentally timed events
tissue-specific and/ or developmental stage-specific expression
can be transported to other cells
if miRNA is poorly regulated what can it lead to
cancer development
what type of RNA plays a critical role in tooth development
miRNA
in what areas of diagnostics are genetics involved in
Polymerase Chain Reaction (PCR)
quantitative reverse transcriptase PCR (qRT-PCR)
whole genome sequencing
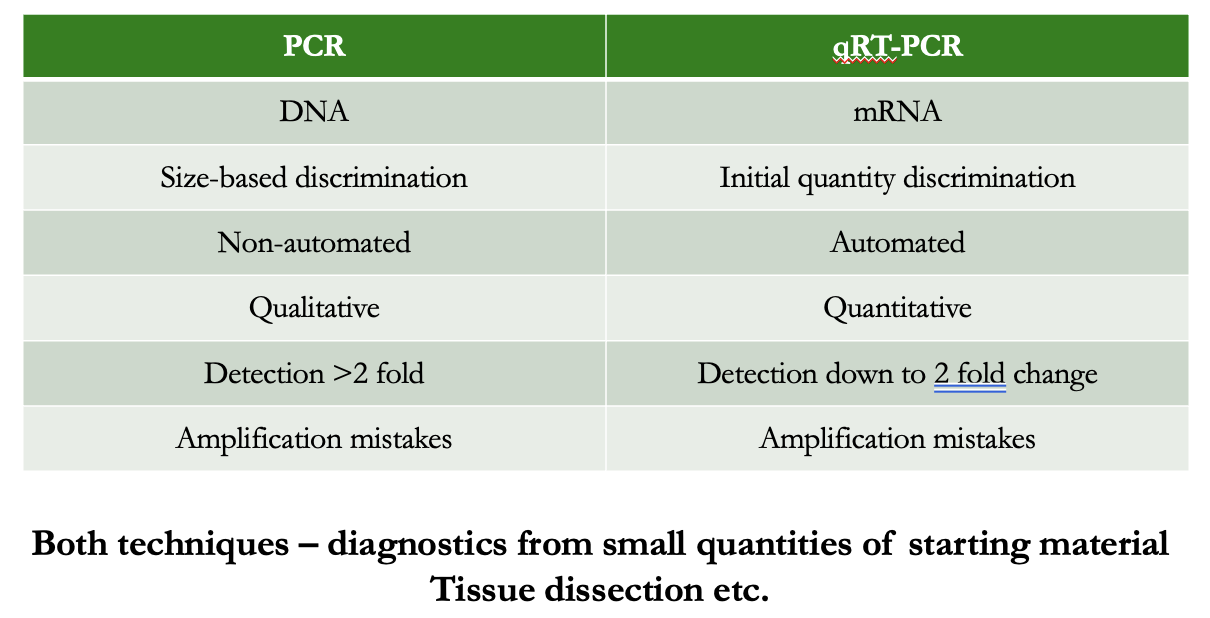
outline PCR
form of DNA amplification
starting material: DNA
amplifies a specific section of DNA
allows visualisation of gene presence or absence on a gel
what does PCR use
thermostable DNA polymerase from bacteria (Thermus aquaticus)
template DNA
primers
dNTPs (A, T, C, G)
Mg2+
outline qRT-PCR
starting material: mRNA
convert to DNA (cDNA)
detects the accumulation of the amplicon during reaction
different amounts of mRNA in starting sample
high amount: amplifies quickly
low amount: takes longer
quantifies amount of mRNA expression
what is qRT-PCR now used for
heavily used during COVID to detect amount of pathogen
now often used for cancer diagnostics in precision medicine
table comparing PCR and qRT-PCR
outline whole genome sequencing
sequences, assembles, analyses human genome
can analyse for presence or absence of genes or specific mutations in (vast computer processing):
neurological disorders
specific cancers
blood disorders
cost and speed of this vastly improved
what is an issue with whole genome sequencing
data storage
outline salivomics
able to monitor (uses computer programming):
genomics, transcriptomics, proteomics, metabolomics, microbiome
what is GWAS
Genome Wide Association Studies
rapid scanning of genomes of many people to find genetic variations associated with a particular disease
precision medicine