RRoy 11: Translation I
1/29
Earn XP
Description and Tags
November 17th, 2025
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
What is mRNA surveillance?
Gatekeeper makes sure incorrect mRNAs are not allowed through the pore complex (quality control on the nucleoplasmic side)
Improperly processed pre-mRNAs are marked for degradation
Incompletely spliced pre-mRNAs that are still associated with the spliceosome components are stopped from leaving
Where are the stop codons in mRNA usually?
They are usually in the last exon
What is nonsense-mediated decay? (NMD)
Once the mRNA gets through the nuclear pore complex, cytoplasmic remodelling takes place (helicase unwinds RNA, cytoplasmic proteins bind, original proteins return to the nucleus)
Every once in a while, the nuclear proteins are improperly removed and remain stuck to the mRNA on the cytoplasmic side
NMD targets mRNA that still have nuclear proteins attached
They detect faulty mRNA by detecting if they have a stop codon before the last exon - This prevents ribosomes from making incomplete RNA as the ribosome will stop at the premature stop codon
What is a dominant negative protein? What causes it?
A dominant negative protein is a mutated protein that affects the function of its non-mutated counterpart (its similarity allows it to block binding sites)
This is caused by mRNA that have an early stop codon (before the last exon)
This is why we need NMD
When is Nonsense-Mediated Decay (NMD) initiated?
It is initiated once mRNA gets into the cytoplasm and ribosomes start to interact with the 5’ end to begin translation
What is the first round of translation called? Why is it called this? What effect does an in-frame stop codon have?
It is called the pioneer round of translation
The first ribosomes that go through the transcript go along the mRNA and remove any nuclear proteins that the helicase/nuclear pore complex missed. This function is unique to the first round
If there is an early stop codon, this is an issue as the pioneer ribosome will detach early and leave nuclear proteins attached
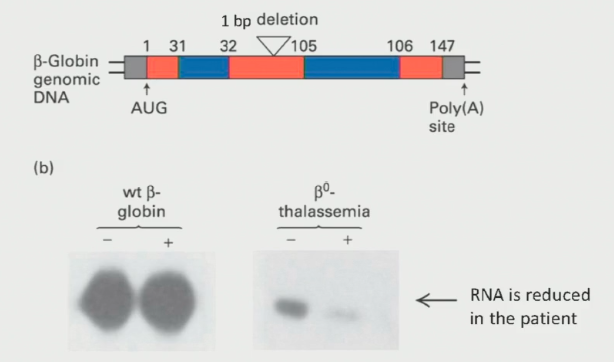
Beta-Thaleassemia Example
In-frame stop example where ribosome detaches early and leaves nuclear proteins attached to distal side of mRNA.
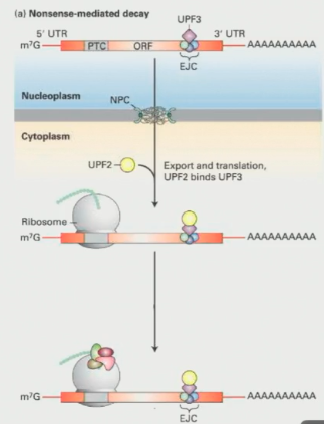
How does Nonsense-Mediated Decay work?
After splicing, a set of proteins called the exon-junction complex (EJC) sits on the exon-exon junction
One exon junction complex, known as Upf3, stays attached to the mRNA as it moves to the cytoplasm
Ribosomes remove Upf3 during the pioneer round of translation. HOWEVER, they cannot displace Upf3 if the stop codon comes before it (defective mRNA)
If Upf3 stays attached due to an early stop, it triggers the formation of a complex that targets the mRNA for degradation (creating a loop-like structure)
When the cell detects this abnormal loop structure, it moves the mRNA to a part of the cell specialized in degradation, known as P-bodies
The three roles of RNA in protein synthesis
mRNA works as a template for the ribosomes, which codes for the specific amino acids
tRNAs attach the corresponding amino acids
rRNA is also critical because they play an important role in assembling the ribosome
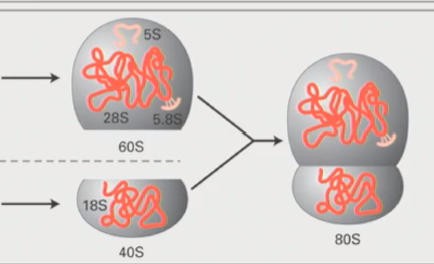
Large ribosomal subunit composition in bacteria
Made up of 23S ribosomal RNA coupled with a 5S RNA (made by RNA polymerase III
Together these combine to form the 50S large subunit

Small ribosomal subunit composition in bacteria
16S ribosomal RNA combines with other proteins to form 30S small subunit

Bacterial ribosome composition
Together the 50S large subunit and the 30S small subunit combine to form the 70S ribosome

Large ribosomal subunit composition in eukaryotes
Combination of 28S and 5.8S ribosomal RNA with 5S to form 60S large subunit

Small ribosomal subunit composition in eukaryotes
18S rRNA compbines with other proteins to form a 40S small ribosomal subunit

Eukaryotic ribosome composition
Large 60S subunit and small 40S subunit combnie to form 80S eukaryotic ribosome
Folding of rRNA (ribosomal RNA)
Ribosomal RNA folds into a complicated secondary structure
This structure is highly conserved
It is critical for catalytic events
Aminoacyl-tRNA synthesases function (hint: where do amino acids attach?)
Catalyze the attachment of amino acids to their corresponding tRNA molecules, ensuring correct translation of mRNA into proteins.
Aminoacyl-tRNAs come in a variety of different flavours, each one carrying a specific amino acid
They can recbognize more than 1 tRNA, since many codon combinations can give rise to the same amino acid
The enzyme has a specific pocket which recognizes it cognate tRNA component, and then hydrolyzes ATP to link the amino acid to the tRNA at the 3’ CCA end.
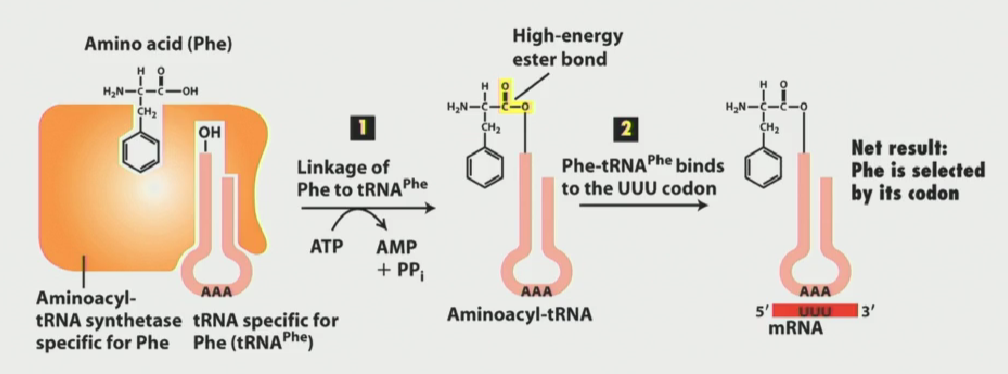
Why is the genetic code considered degenerate?
A single aminoacyl tRNA synthetase can bind to more than one unique tRNA
A single codon can be bound by more than one single tRNA (anticodon)
Two types of methionine tRNA (when are they used?)
One tRNA met is used to code for methionine in the elongation phase of protein synthesis
Other tRNA met is used for initiation only (written as tRNAi)
Both tRNA mets are charged by the same aminoacyl tRNA synthetase
in bacteria, the initiator tRNAi met is modified (a formyl group gets added) to distinguish it from normal tRNA met
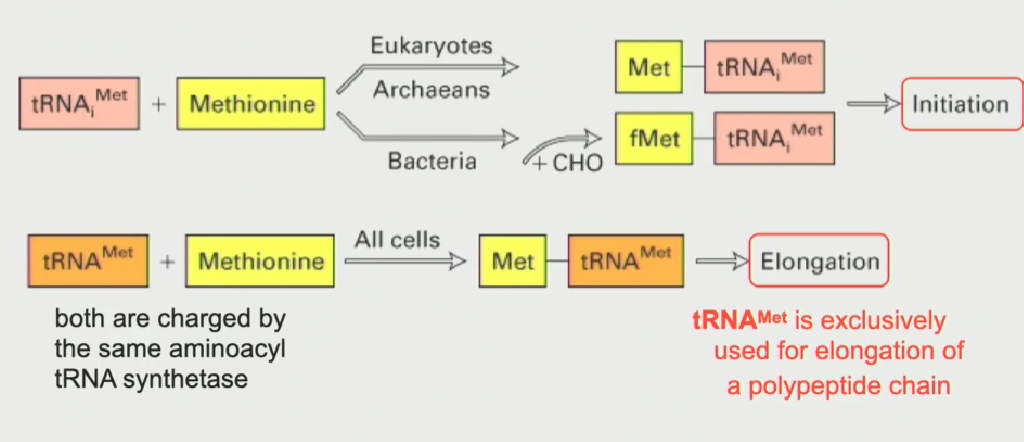
Unique structural function of tRNAi met
Its' structure allows it to interact with a specific site on the small ribosomal subunit (known as the P-site)
This is why only this tRNA is used for initiation of a sequence
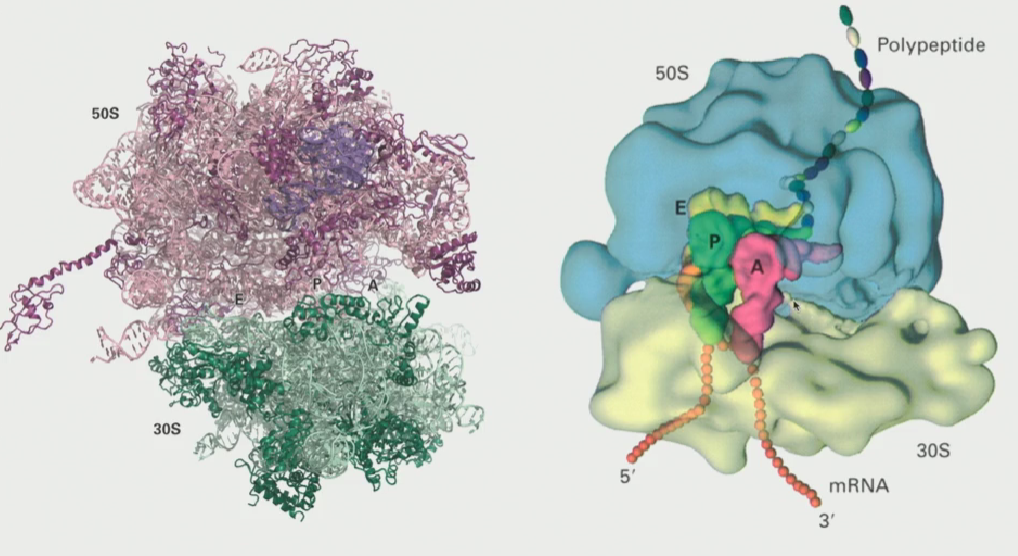
3 sites in the ribosome
A site (acceptor)
P site (polypeptide)
E site (ejector)
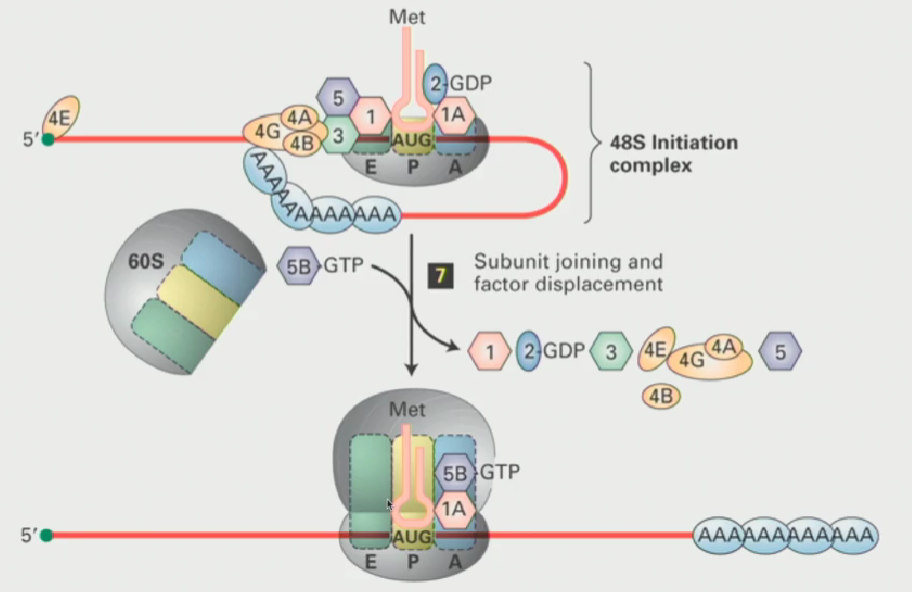
Assembly of the pre-initiation complex
When the ribosomes are not translating, they subunits have to be kept apart
The small ribosomal subunit interacts with proteins called initiation factors (or eukaryotic initiation factors)
These proteins ensure that the two subunits do not come together unless triggered
Small ribosomal subunit has a free P-site
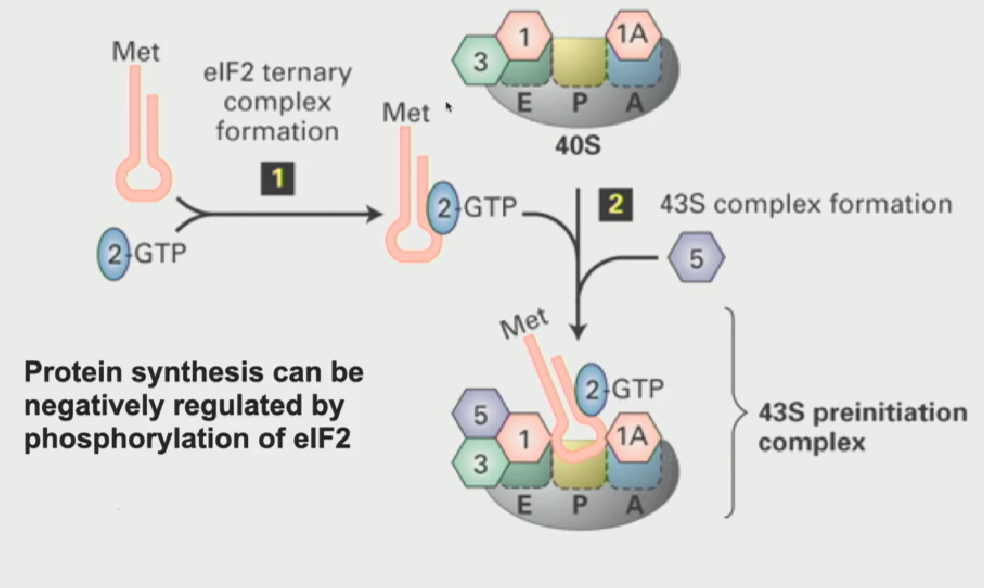
Another eukaryotic initiation factor known as eIF2 (a GTP binding protein), when active, interacts with the tRNAi met (initiator tRNA), causing it to bind to the P-site on the small ribosomal subunit
This results in a 43S pre-initiation complex (small ribosomal subunit is ready to translate)
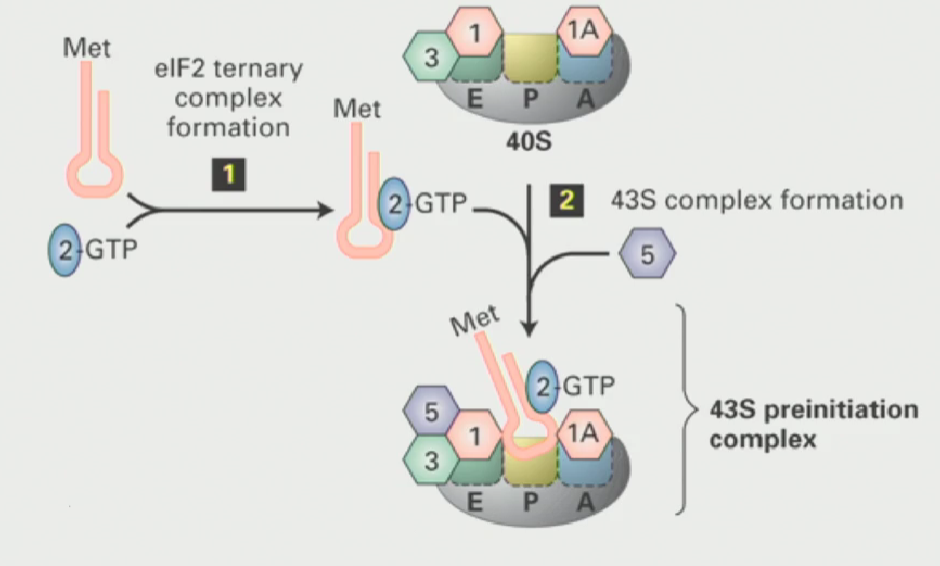
Regulation of the 43S pre-initiation complex
highly regulated process
When growing conditions are very poor (ex. periods of starvation), you do not want to translate proteins. So, you can block all protein synthesis by phosphorylating eIF2
Once it is phosphorylated, it can never bind GTP to become activated → cannot bind tRNAi met (form ternary complex) → 43S preinitiation complex does not form → small ribosomal subunit doesnt translate
Function of cap-binding protein (Sonenberg experiment)
the mRNA that is going to be translated has a cap binding protein (eIF4E) at the 5’ end
If you add the cap-binding protein to an in-vitro translation reaction with mRNAs that come from various viruses that are capped and non-capped, the capped mRNAs had enhanced translation. It did not work for uncapped mRNAs
This protein somehow interacts with the cap to enhance protein synthesis from that mRNA
What kind of mRNA is the cap attached to? What kind of cap is it? What is the relation to CDK7?
the 7’methylguanylate cap is only attached to Class II mRNAs (synthesized by RNA polymerase II)
This is because those transcripts come from an RNA polymerase II that is phosphorylated by CDK7 (Recall: on the serine 5 of the CTD heptapeptide repeat)
The capping enzyme needs that serine 5 phosphorylation to use as a platform, to bind with that region where the 5’ end is coming out
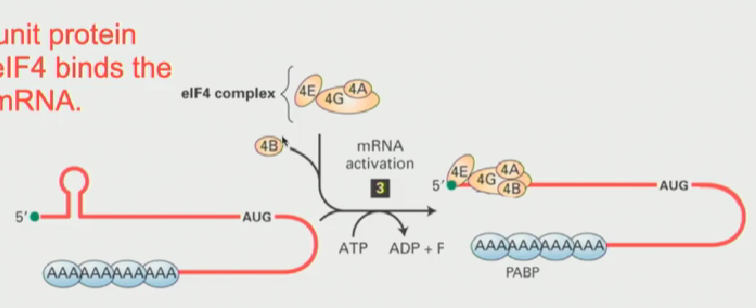
What is eIF4E? What complex was it a part of?
“eukaryotic Initiation Factor 4E”
The eukaryotic capping protein
Part of a big protein complex known as eIF4F
What is eIF4F? What is included?
A collection of smaller subunits that include the cap-binding eIF4E, eIF4A, eIF4B and eIF4G
cap binding protein is critical as it associates with the 5’ cap, indicating that that mRNA is ready to be be translated
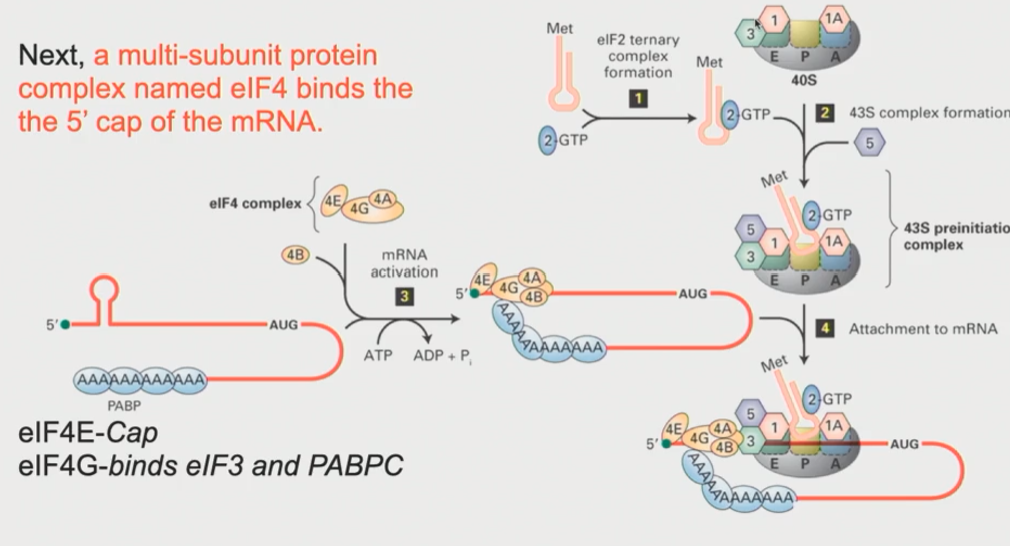
How does the eIF4F complex work? What structure does it form in the mRNA
eIF4E and associated proteins will sit down at the 5’ end of that mRNA
eIF4G acts as an intermediary. Through protein-protein interactions, it interacts with eIF3 on the 43S pre-initiation complex
eIF4G also interacts with the Poly-A tail, through interactions with PABPC1
This makes the mRNA form a loop, where the 3’ end (the Poly-A tail) somehow has to interact with the proteins at the 5’ end (The 43S pre-initiation complex is there)
At this point, the ATP dependent RNA helicase, which is intrinsic to eIF4A, kicks in & the complex begins to scan!
Scanning will ctoninue until a start codon is reached
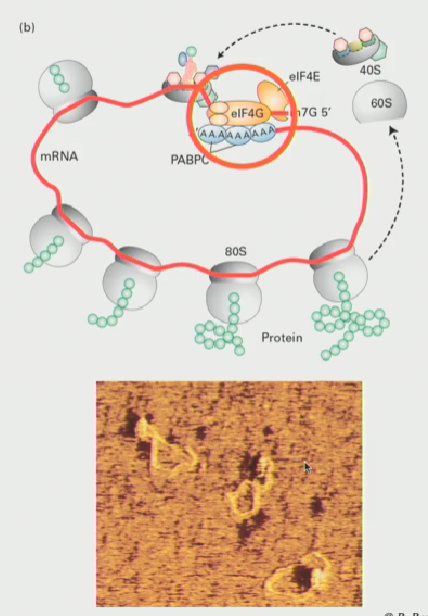
What are the functions of the interactions between the 5’ and 3’ ends of mRNA (aka the loop structure formed by the eIF4F complex
As ribosomes wheel around, eventually it will come to a stop then be ready to translate again
In other words, these loops favour the re-initiation of translation
What happens when the scanning complex arrives at the translational start?
The recognition of the AUG codon by tRNAi met leads to hydrolysis of the GTP associated with eIF2 (making it inactive) and, following that, the initiation factors dissociate
Then, eIF5B in a GTP bound form interactswith eIF1A to create the active ribosome
The eIF5B GTP is hydrolyzed and other initiation factors will release, now the ribosome is ready to transcribe