DNA Polymerase & Replication
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
38 Terms
Kornberg Experiments: Discovery of RNA Polymerase
grow E.coli (we know we that E.coli has everything it needs to replicate DNA; so the polymerase must be in there)
break open cells
prepare a soluble extract
fractionate extract to resolve different proteins (and repeat)
look for incorporation of radioactivity in polymerized DNA (which has a different solubility than free nucleotides)
Purification Strategy
take a complex mixture → cells are broken open → membrane components spun down thru centrifugation
left w/ soluble fraction (many diff proteins in this mixture)
now can be separated based on different biophysical properties (e.g size, hydrophobicity, charge)
purifications: size, solubility, surface charge, binding capacity, etc.
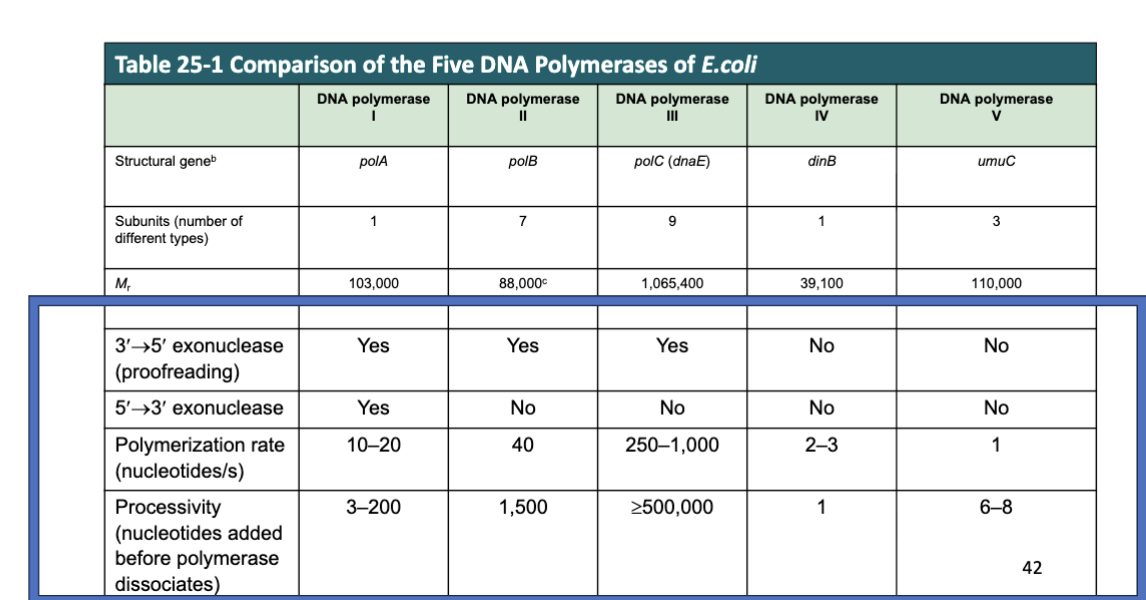
Kornberg Activity Assay
used radioactive thymidine b/c it is found in DNA, but not RNA → allowed them to isolate DNA polymerase
checking whether any mixtures allow for the synthesis of a polymer of labeled thymidine into DNA
need to separate the labeled nucleotide from polymerized DNA
after reaction: added acid to DNA → polymerized DNA precipitates
unincorporated labeled nucleotide would remain free in solution
radioactivity in the insoluble fraction indicates DNA synthesis
DNase breaks up polymers of DNA into single nucleotide
does it switch from pellet into soluble fraction
Kornberg Activity Assay FIGURE
Kornberg Activity Assay: Next Steps
rxns confirmed presence of protein by DNAase (still very many) → repeat steps (eg. fractionation) until they can isolate a homogenous solution of DNA polymers
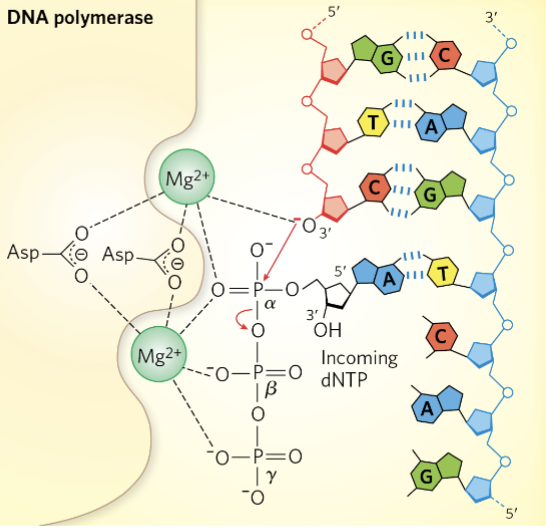
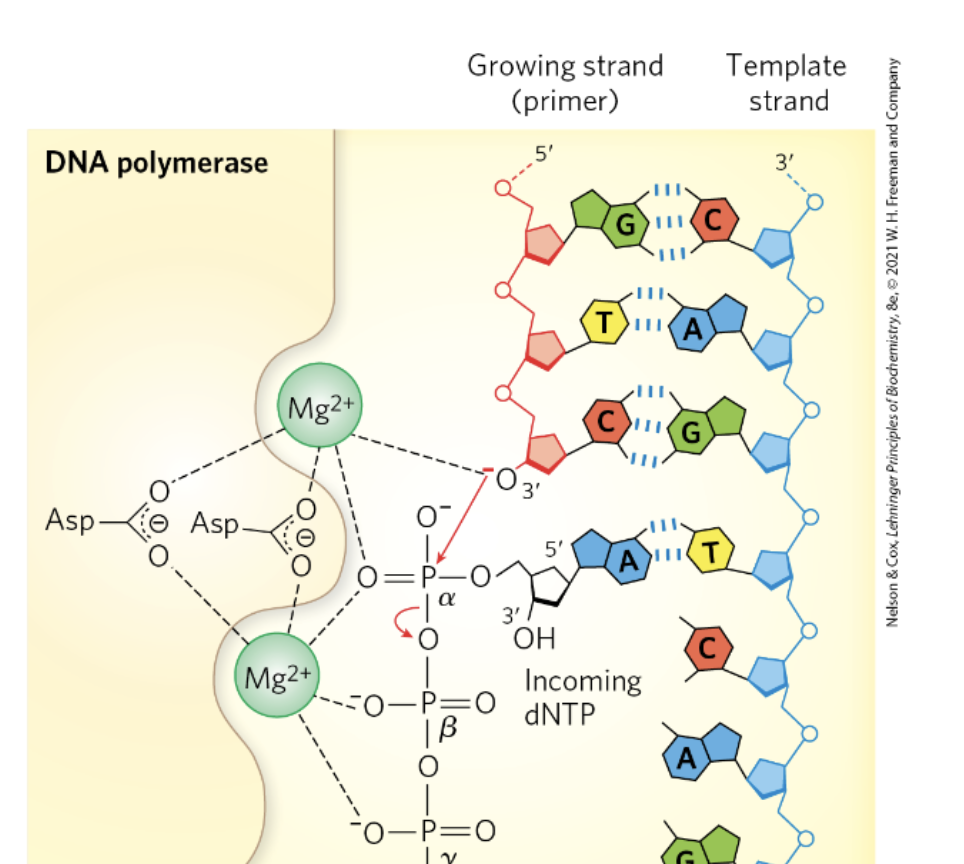
DNA Polymerase Reaction Mechanism
incoming (antiparallel) triphosphate nucleotide forms W-C (canonical interaction) w/ template strand
phosphodiester bond b/w 3’OH of most recently added base and ⍺-phosphate of incoming nucleotide
magnesium stabilizes 3’OH and ⍺-phosphate (intermediate state; temporary (-) charge on 3’OH; allows for nucleophilic attack)
β and γ phosphate is released as PPi new phosphodiester bond is formed
as long as template strand is still available a new substrate is generated for another round of polymerization
DNA Polymerase Mechanism Figure
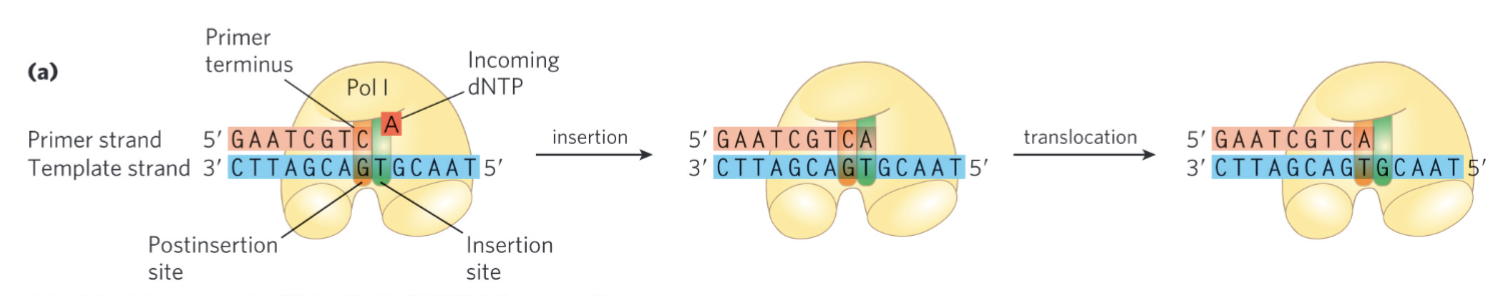
Requirement of DNA-templated DNA polymerase
single-stranded template
deoxyribonucleotides w/ 5’ triphosphate (dNTPs)
Mg2+ ions (essential co-factor for polymerase): coordinate the reaction and neutralize (-) charge
annealed primer (often RNA) w/ a free 3’OH: or else can’t make phosphodiester bond and no new nucleotides can be added
the free 3’OH is necessary b/c of 5’→3’ directionality
Single-Stranded Template
polymerization is guided by a template DNA strand according to W-C base pairing rules
first step is template guided; ensures we’re getting a correct copy during replication (b/c antiparallel complementarity is preserved)
single-stranded template in vivo is generated by a helicase (ATP-dependent specialized protein that unwinds double-stranded nucleic acid polymers)
Single-stranded Binding Protein
once unwound, single-stranded DNA is protected by single-stranded binding protein
temporarily binds binding protein
requirements to work:
needs to be able to interact w/ any type of single-stranded DNA (can’t be sequence-specific)
must easily slide out the way (DNA replication is very fast)
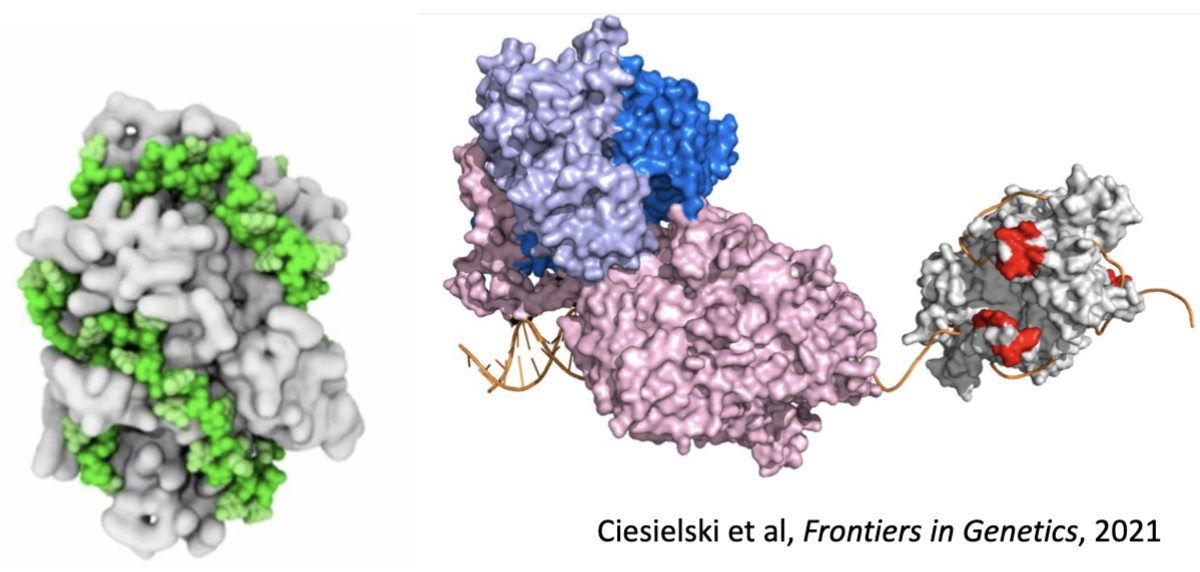
Contribution of Base-Pair Geometry to Fidelity of DNA Replication
Shape discrimination
formation of W-C interactions: highly favorable b/c of complementary H-bonding capacity + active site that fits W-C interactions perfectly
the active site has enough flexibility to enable catalysis of properly matched bases
incorrect base pairing results in contortions of the binding pocket to form H-bonds b/w the template and the incoming nucleotide (much less favorable)
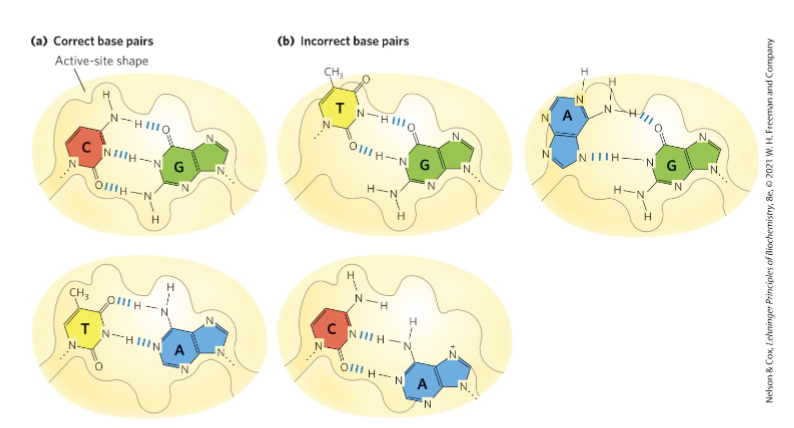
Why is replication accurate?
formation of H-bonds
geometry of the active site
exonuclease editing capcity
mismatch repair
Why replication is possible
DNA must be replicated every cell cycle
its double-stranded complementary structure suggests a copying mechanism
unraveling the two parental strands results in the production of two identical DNA molecules
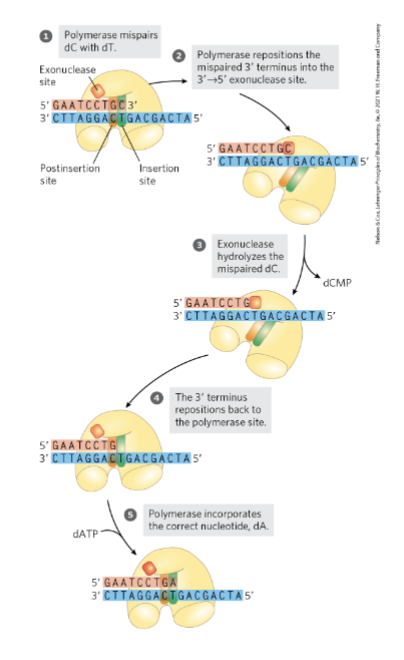
Error Correction by DNA Polymerase I
translocation of the enzyme is inhibited when an incorrect nucleotide is added
makes non W-C interactions w/ template
phosphodiester backbone is formed
doesn’t fit in active site
many DNA polymerases have intrinsic 3’→5’ exonuclease proofreading activity
permits the enzyme to remove a newly added nucleotide
mismatched pairs fit well in the editing site (more favorable)
makes a break in the phosphodiester bond
Proposed Models of DNA Replication
Conservative: replication of both strands, but parental strands would come tgt and daughter strands come tgt
Semi-conservative: new daughter strand stays attached to parental strand (2 new strands formed from 1 daughter + 1 parent strand)
Dispersive: DNA is broken up → replication → reanneal back tgt

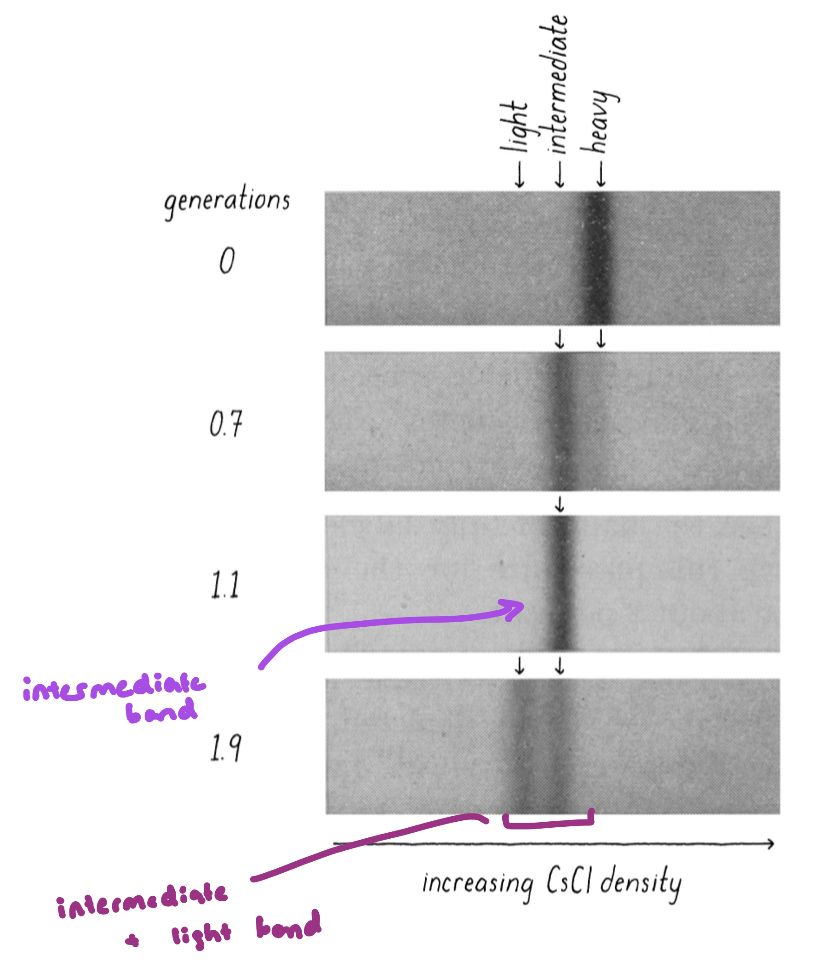
Meselson-Stahl Experiment
grew E.coli in the presence of heavy nitrogen (15N) to label the bacteria’s DNA
grew for many generations → know all the genome is heavy
bacteria copy their entire complement of DNA, or genome, before every cell division
then moved the bacteria to a normal 14N-containing medium
allowed the cells to divide once
separated the DNA by density
the results supported semi-conservative replication
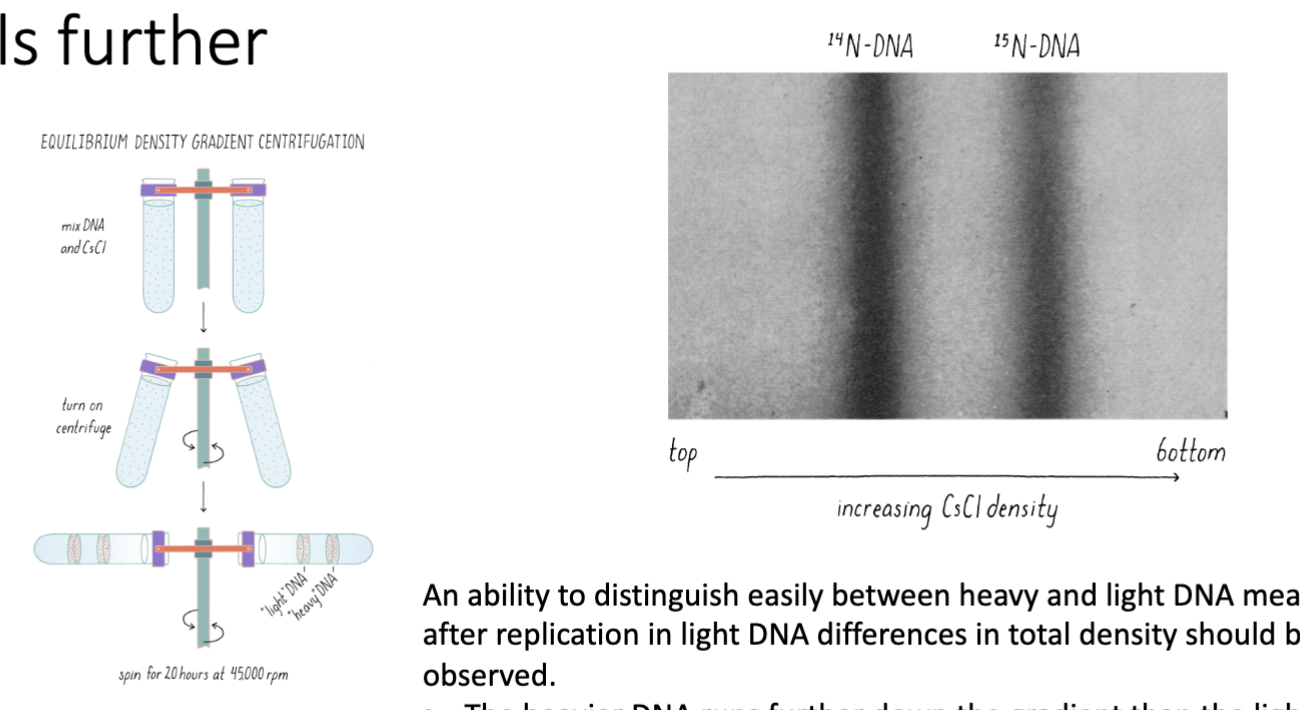
Heavy vs Light DNA on Cesium Chloride Density Gradient
heavier DNA travels further
an ability to distinguish easily b/w heavy and light DNA means that after replication in light DNA differences in total density should be observed
Meselson-Stahl Experiment: Semi-convservative replication results
after 1 generation (1 doubling of the E. coli – means all the DNA has been duplicated) the DNA from these samples is a mixture of heavy and light (intermediate density)
rules out conservative replication mode
after 2 generations, the DNA is either all light, or a mixture of heavy and light
supports the semi conservative model
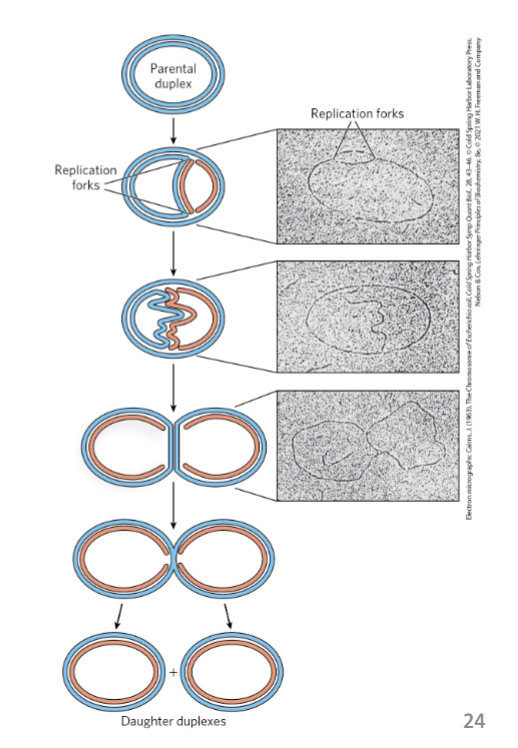
Bidirectional Replication
replication begins at an origin and proceeds bidirectionally
replication forks = dynamic points where parent DNA is being unwound and separated strands replicated
both DNA strands are replicated simultaneously
both ends of the bacterial chromosome have active replication forks (bidirectional replication)
Primase
adds RNA primer that DNA polymerase can use as a substrate
short DNA sequence (RNA polymerase-like rxn is mechanistically similar)
1st nucleotide (has 5’ phosphate) added just by making W-C interactions
RNA primer can’t be added by DNA polymerase; required RNA polymer-like enzyme (in E.coli, added by pol-⍺)
Primase Figure
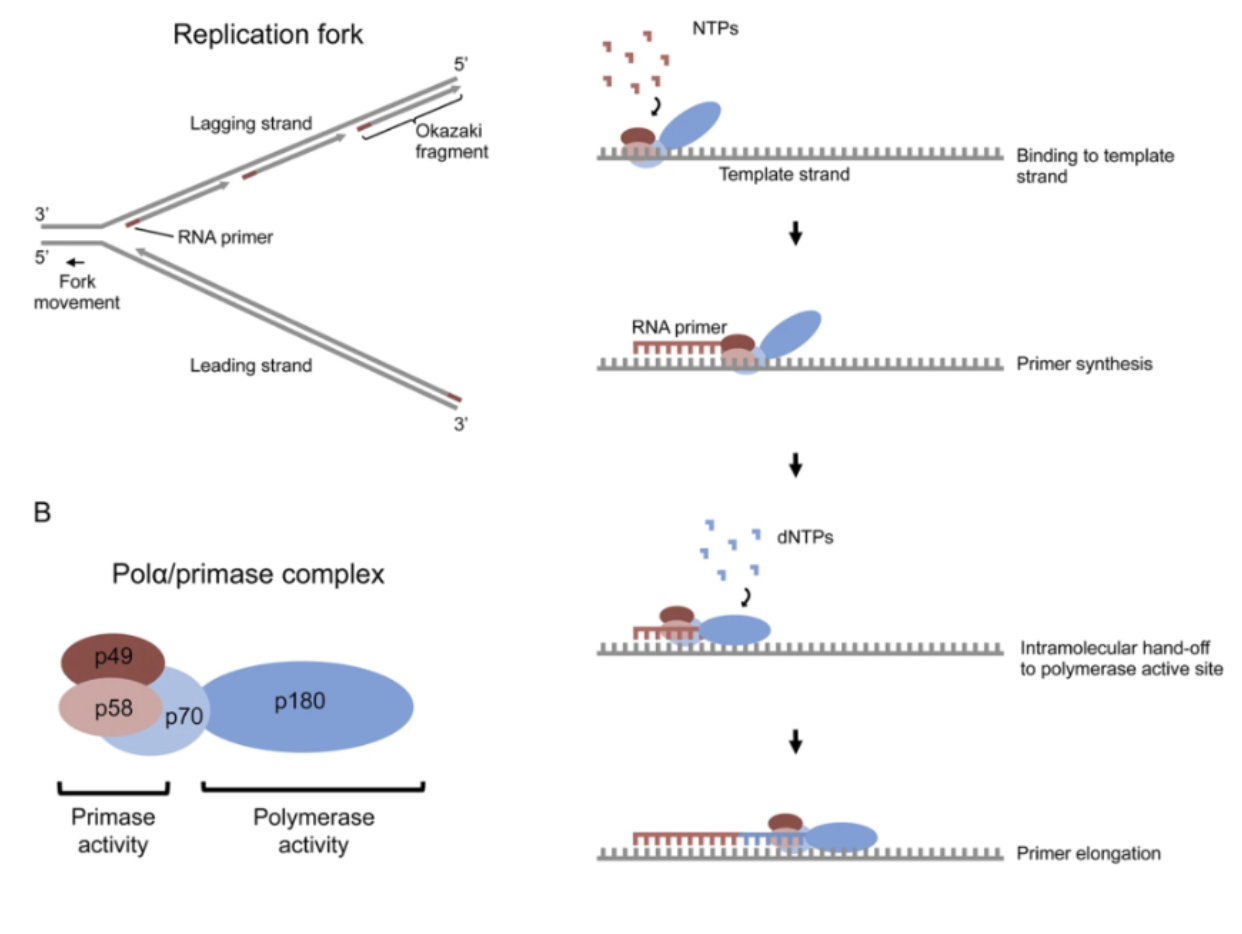
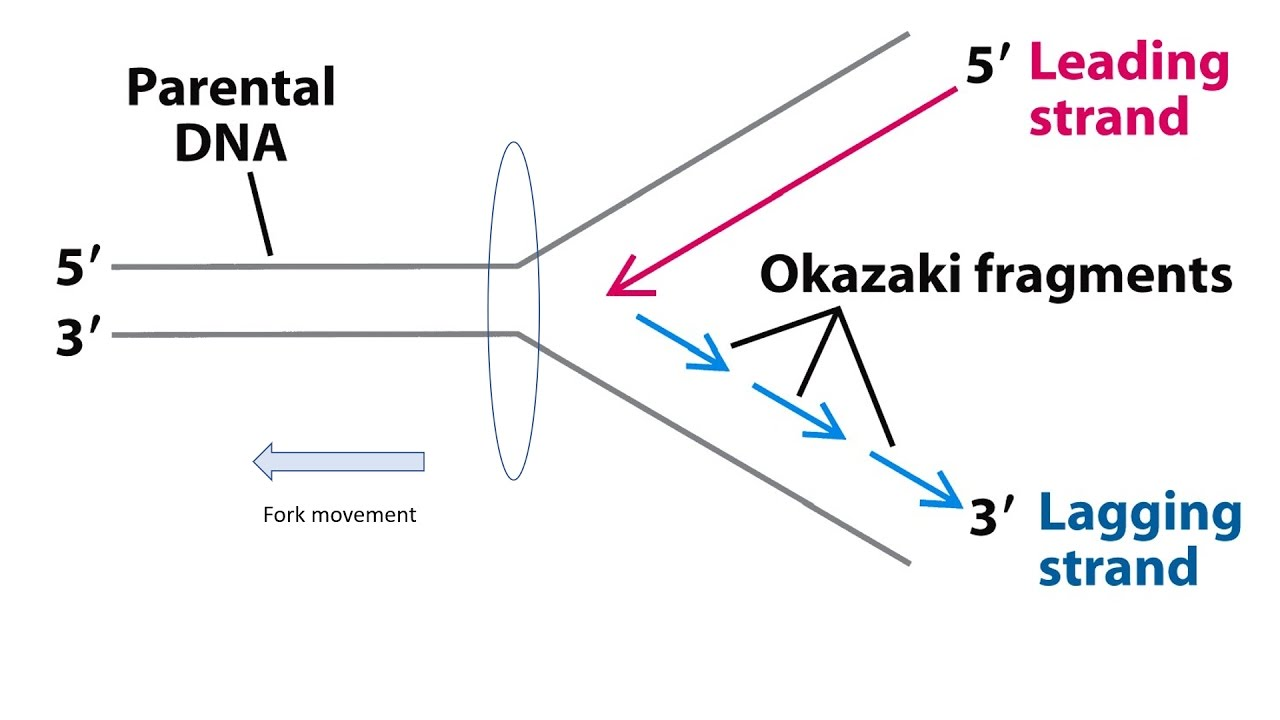
Okazaki Fragments
short DNA segments created on the lagging strand during DNA replication
lagging strands synthesis requires discontinuous, piece-by-piece production that DNA polymerase then joins tgt w/ DNA ligase
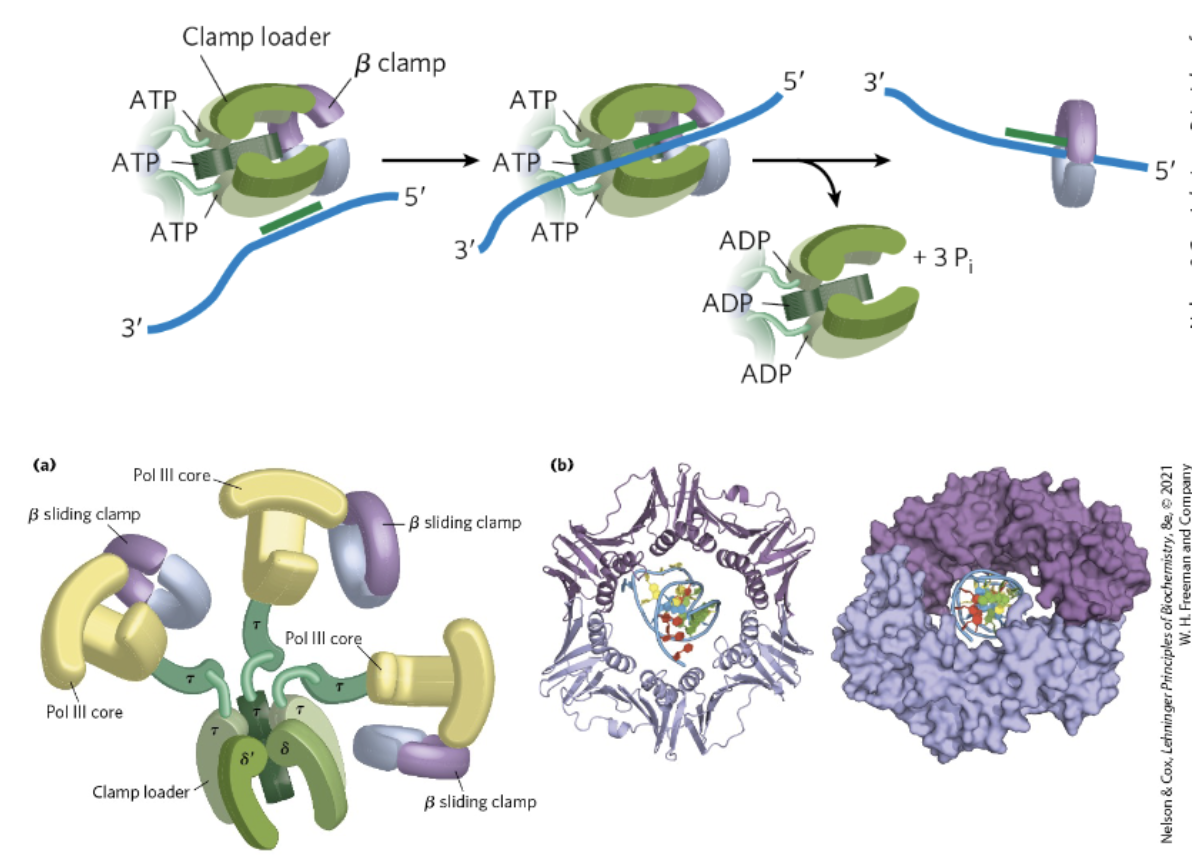
DNA Polymerase III Clamp Loader
clamp is loaded onto the DNA-RNA hybrid by a clamp loader in ATP dependent rxn
prevents DNA polymerase from falling off
uses power of ATP hydrolysis to provide mechanical force so that it’s able to open up the ring structure and latch it down
clamp loading requires ATP binding and hydrolysis to add the clamp to the substrate
latches over RNA/DNA hybrid structure formed after primase adds RNA primer
going over a double-stranded region
clamps onto the back of polymerase and follow along behind polymerase
DNA Polymerase III Clamp Loader FIGURE
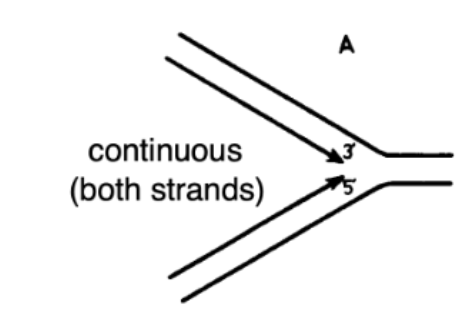
Replication is Semi-Discontinuous: Continuous Hypothesis
Not true
missing a polymerase that could add free nucleotides to the 5’ end and grow strand in opposite direction
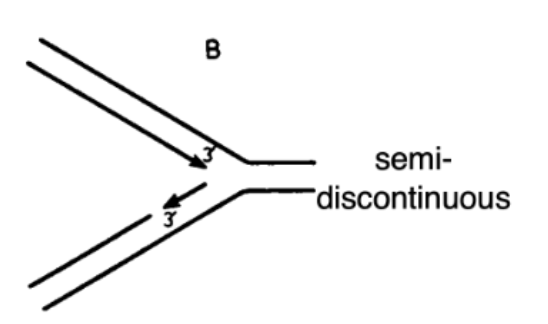
Replication is Semi-Discontinuous: Semi-Continuous Hypothesis
True
one strand primed at one end and then goes all the way until end of linear chromosome
other strand is synthesized in pieces
enough single-stranded region has to be pulled out fork and for primase
Processivity
how many nucleotides is DNA polymerase able to add before falling off
Leading Strand Synthesis
helicase (DnaB) unwinds DNA at the replication fork
primase adds a primer to generate a free 3’OH
clam loader loads on a clamp to ensure synthesis is pocessive
DNA polymerase synthesizes the leading strand is synthesized in one piece in 5’→3’ directions
follows helicase until end of sequence
Lagging Strand Synthesis
synthesized in Okazaki fragments
primase repeatedly adds primers to generate new 3’OH groups
clamp loader repeatedly loads sliding clamps onto new RNA-DNA primer template hybrids
DNA polymerase extends each fragment until it reaches the primer of the previous Okazaki fragment
cycle repeats until lagging strand synthesis is complete
Lagging Strand Synthesis (Slide notes)
trombone mechanism
lagging strand synthesis required loop formation
single strand at fork comes out far enough for primase to bind and add RNA primer (w/ free 3’OH)
happens in opposite direction of fork
once no more single-stranded synthesis → falls off
process is repeated → generates fragments as replication process occurs
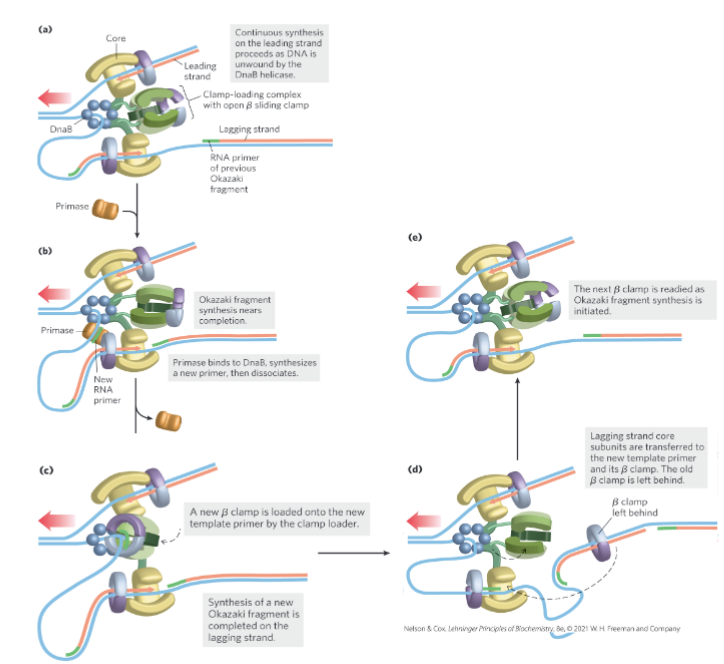
RNA and Gaps Problem
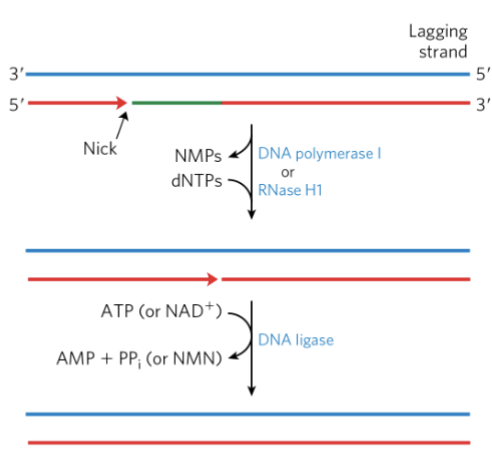
synthesis of short fragments of DNA generates two new problems
RNA problem → polymerase can’t fill the gap (needs 3’OH)
once the last nucleotide is added, can’t generate the last phosphodiester bonds (done by DNA ligase)
generating Okazaki fragments involves short regions of RNA-DNA hybrid complexes in the lagging strand
lagging strand contains many RNA primers, leading strand = 1 RNA fragment
we don’t want RNA in our genome; has to be removed
DNA pol I removes the RNA and replaces it w/ DNA thru the specialized exonuclease activity (nick translation)
DNA ligase seals the remaining nick
RNA and Gaps Problem Figure
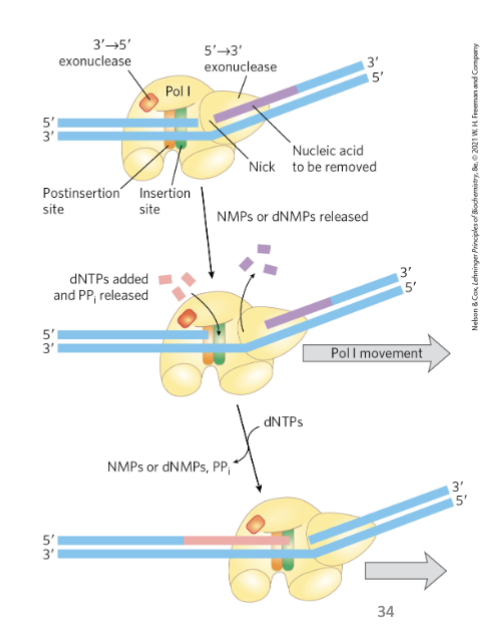
DNA Polymerase I: 5’→3’ Exonuclease Activity
distinct from 3’→5’ proofreading exonuclease
the 5’→3’ domain is in front of the enzyme and performs nick translation
removes RNA in the 5’→3’ direction
mild protease treatment separates this domain from the remainder of the enzyme (the large fragment or Klenow fragment which is responsible for the nick translation function of DNA pol I)
Nick Translation
occurs in the 5’→3’ exonuclease domain
RNA removed one at a a time, and dNTPs are added as RNA is removed (same active site)
nick translation = a break or nick in the DNA is moved along with the enzyme
important in: DNA repair, and removal of RNA primers during replication
Proteins acting at the Replication Fork
Proteins that are required to replicate genome during s-phase; DNA polymerase cannot replicate the entire genome, several other proteins are required
SSB: binds and stabilizes single-stranded DNA generated by helicase
helicase: DNA unwinding (polymerase only works w/ single-stranded template
primase: RNA primer synthesis
DNA pol III: new strand elongation (E. coli only has this)
DNA pol I (pol ⍺): filling of gaps; excision of primers
DNA ligase: ligation
DNA gyrase (topoisomerase II): supercoiling; reduces strain
pushing polymerase thru a genome (especially circular) results in lots of (+) supercoiling → strain/stress
More than on DNA polymerase (Paula DeLucia)
DeLucia and Cairns isolated a mutant E.coli w/ no DNA pol I activity but could still survive
the polymerase responsible for replicating E.coli chromosome in vivo is DNA pol III (discovered by Kornberg)
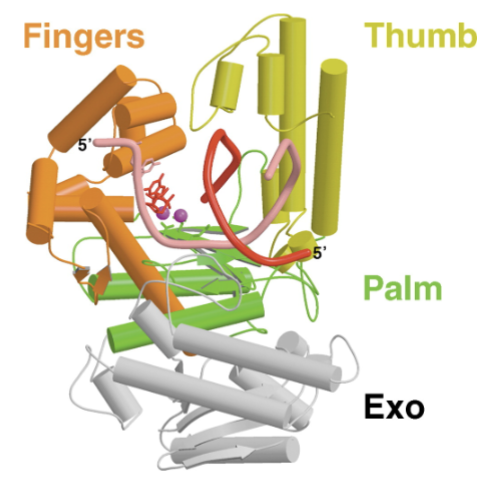
DNA pol I is the prototype for all DNA polymerases (enzymatically and structurally)
the catalytic site and overall fold (hand structure) is conserved, editing capacity varies depending on the polymerase
E. coli has at least 5 DNA polymerases
DNA polymerase I is abundant, but insufficient for replication for the E. coli chromosome
rate (600 nucleotides/min) is slower than observed for replication fork movement
low processivity
the primary function of DNA pol I is cleanup during replication, recombination and repair
DNA Pol II, III, IV. and V
DNA pol I: involved in RNA removal post-replication and DNA repair
DNA pol II: involved in DNA repair
DNA pol III: the principle replication enzyme in E. coli
IV and V: involved in an unusual form of DNA repair
translesion polymerases that repair damage from UV radiation