gene regulation - prokaryotes
1/12
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
13 Terms
the genotype and phenotypes of bacteria
The potential phenotypes of bacteria is carried in their DNA
The genotype is the potential to show different features
Phenotype is the expression of genetic traits in the genotype
The features shown by the bacteria are their phenotype
Phenotypes change depending on the environment
the bacterial genome
The majority of bacterial genomes are circular (Streptomyces) are linear
Linear in size
Extra-chromosomal DNA & plasmids give bacteria extra properties
Some associated with antibiotic resistance
Plasmids can be transferred between bacterial cells (horizontal gene transfer)
variation in bacterial genomes
Vary in size and number
Generally, a linear relationship between genome size and number of genes
Genes tightly packed
Almost no introns
Little space between gene coding regions
On average one open reading frame (ORF) per 1-1.2 kb
On average 20-40% of every bacterial genome is unique to that organism
Functions of these genes are often unknown
how to create a phenotype
Bacteria sense their environment to ensure their 'transcriptome' is optimal for their environment
There are 3 major stages of transcription, all of which can be regulated
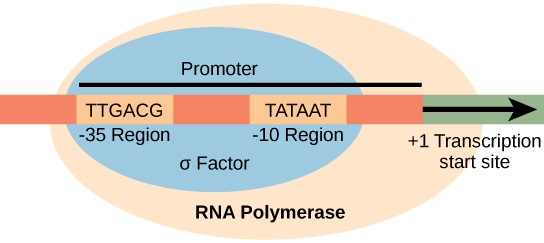
Initiation
Requires a sigma factor that recognises the –10 and –35 region
It allows RNA polymerase to know where to initiate transcription
Opens DNA to enable transcription
Housekeeping sigma factor (transcription of key genes for bacterial cell function)
There are alternative sigma factors (e.g. stress responses)
Elongation
Not at a uniform speed
Speed and pausing is determined by the DNA sequence being transcribed and RNA secondary structure
Proteins can be bound to the DNA to block transcription
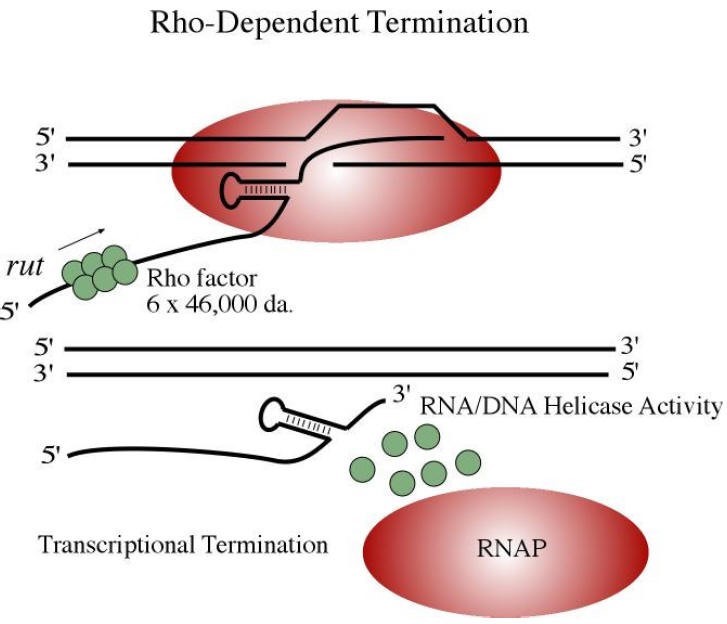
Termination
Terminators are punctuation marks within the DNA sequence, defining where transcription stops
Is intrinsic
Rho dependent termination
Transcription is catalysed by a single RNAP
RNAP core enzyme is composed of five sub-units
2 x beta
2 x alpha
1 x omega
bacterial messenger RNAs
No nucleus through which mRNAs need to be transported
Effectively no processing of mRNAs before translation
Do not have methyl-G caps or poly-A tails
Do have NAD capping
Newly made (nascent) mRNA transcripts are translated
Thus bacteria are able to rapidly synthesise proteins
Relatively less stable than eukaryotic mRNA
Half-life of bacterial mRNA is roughly 2 mins
It is 4-24 hours for eukaryotes
transcription factors
Turn gene expression on or off
Bind to DNA
Normally near the start of the transcriptional start site
Recognise specific sequences in the DNA
Gene expression specificity
Either help or hinder binding of RNA polymerase
Respond to environmental signals
Positive control (with activator proteins)
Negative control (with repressor proteins)
Negative repressible operons
transcription factors
positive control - with activator proteins
RNA polymerase can only bind to the promoter region if an activator protein binds to a site near the promoter
If the activator does not bind to the DNA, the RNA polymerase cannot bind and transcription does not occur
transcription factors
negative control - with repressor proteins
The natural state of the DNA allows the RNA polymerase complex to be recruited and transcription takes place
If the repressor protein binds to the DNA, it inhibits recruitment of RNA polymerase and transcription does not occur
transcription factors
negative repressible operons
On until turned off
No corepressor
Repressor is inactive
Transcription occurs
Corepressor present
Repressor binds to corepressor
Causes conformational change in shape
Forms an active complex
Transcription is blocked
activation of transcription - LuxR
Quorum sensing – molecular system to monitor population density
Bacteria produce autoinducer molecules
Can be a peptide or a small molecule
Concentration dependent
Linked to density of the population
Population becomes quorate to co-ordinate changes in behaviour
This is recognised by a transcription factor – LuxR
In the absence of the molecule, the DNA binding site is revealed and LuxR can recognise transcription factor binding sites
Transcription of gene
tryptophan repression of transcription
TetR acts as a transcriptional repressor
Binds to tetO boxes (specific sequences)
Blocks interaction of RNA polymerase
When tetracycline is added binds to TetR
Causes conformational change in shape so can no longer bind to tetO
RNA polymerase binds
repression of transcription
controlling iron levels
70% of iron in the human body is in the form of heme
In other environments iron is a rare nutrient
Essential for many important biological processes
Too much iron can be dangerous for cells
Causes DNA damage due to oxidative stress
Fur is a transcription factor that detects iron availability
Binds to Fe2+
When iron binds to Fur
Complex binds to Fur box
No transcription
Happens in an iron abundant environment
When iron does not bind to Fur
Not complimentary to Fur box
RNA polymerase binds to DNA
Transcription occurs
Fur regulates Shiga Toxin production
Shiga toxin is produced in the intestinal tract
Oxygen availability is variable, leading to changes in iron availability
If oxygen is absent, Fe2+ is present
If oxygen is present, iron is oxidised to fe3+
Fur is bound to, and transcription occurs
antisense small RNA (sRNA)
Regulatory RNA, complementary to specific mRNA, used to reduce translation