A Level CIE Biology: 16 Inheritance
1/110
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
111 Terms
diploid
cell that contains 2 complete sets of chromosomes (2n) which contain the dna necessary for protein synthesis and cell function.
how mnay cells in body are diploid and how many chromosomes
nearly all with 23 pairs (46 individual) of chromosomes in nucleus
haploid cells
contain one complete set of chromosomes (n), have half no. chromosomes compared to diploid cells. called gametes and involved in sexual reproduction for humans they are female egg and male sperm
how many chromosomes have haploid
cells that contain 23 chromosomes (no pairs) in their nucleus.
haploidy and diploidy
terms can be applied to cells across diff species describe no. sets of chromosomes not total no.

what happens during fertilisation, why are chromosomes important
nuclei of gametes fuse together to form the nucleus of zygote (both gametes must contain correct no. chromosomes for zygote to be viable). if a zygote has too many or too few may not survive.
for a diploid zygote, gametes must be..
haploid. n+n=2n (n is haploid no., 2n is diploid)
what does meiosis do
produces haploid gametes during sexual reproduction
what is the first cell division of meiosis and what happens
reduction division
nuclear division that reduces chromosomes no. cell
in humans 46 diploid → 23 haploid ensuring that gametes are haploid

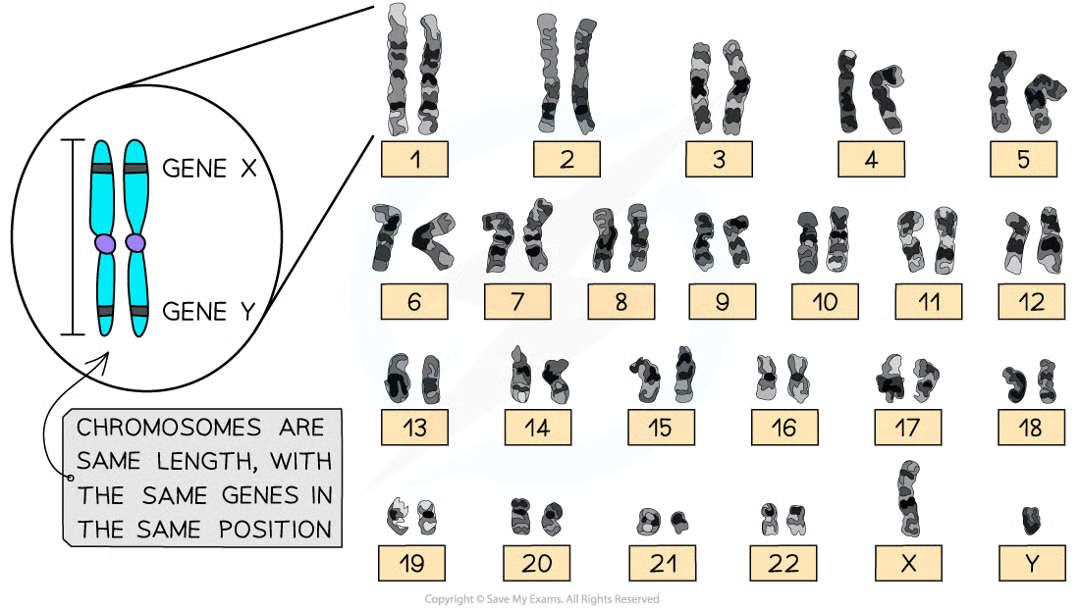
chromosomes characteristic shape:
fixed length, particular location of position of centromere
why are chromosome characteristics important
allow for each chromosomes to be identified in a photomicrograph (chromosmes often grouped into homologous pairs)
homologous chromsomes 2 features
carry same genes in same positions (locus - specific linear position of particular gene on certain chromosome)
same shape
during fert what is formed and how
zygote. one chromsomes of each homologous pair comes from female gamete and other comes from male. same genes in same order helps them line up alongisde each other during meiosis
what is meiosis
a form of nuclear division that results in the production fo haploid cells from diploid cells, producing gametes in plants and animals that are used in sexual reproduction. consists of meiosis I and II (pmat)
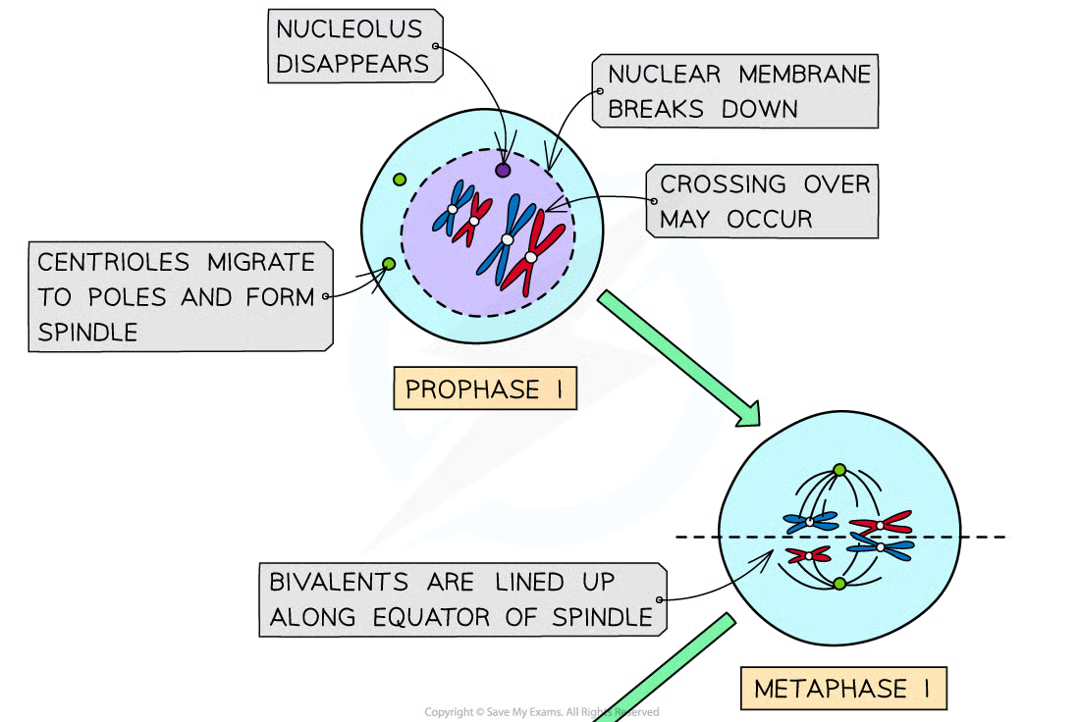
prophase I (6)
dna condenses and becomes visible as chromosomes
dna replication has already occured so each chromosome consists of 2 sister chromatids joined together bya centromere
chromosomes are arranged side by side in homologous pairs (pair = bivalent)
as homologous chromosomes are very close together the crossing over of non-sister chromatids may occur. point at whcihc crossing over occurs = chiasma
centrioles migrate to opposite poles and spindle is formed
nuclear envelope breaks dwon and nucleolus disintegrates
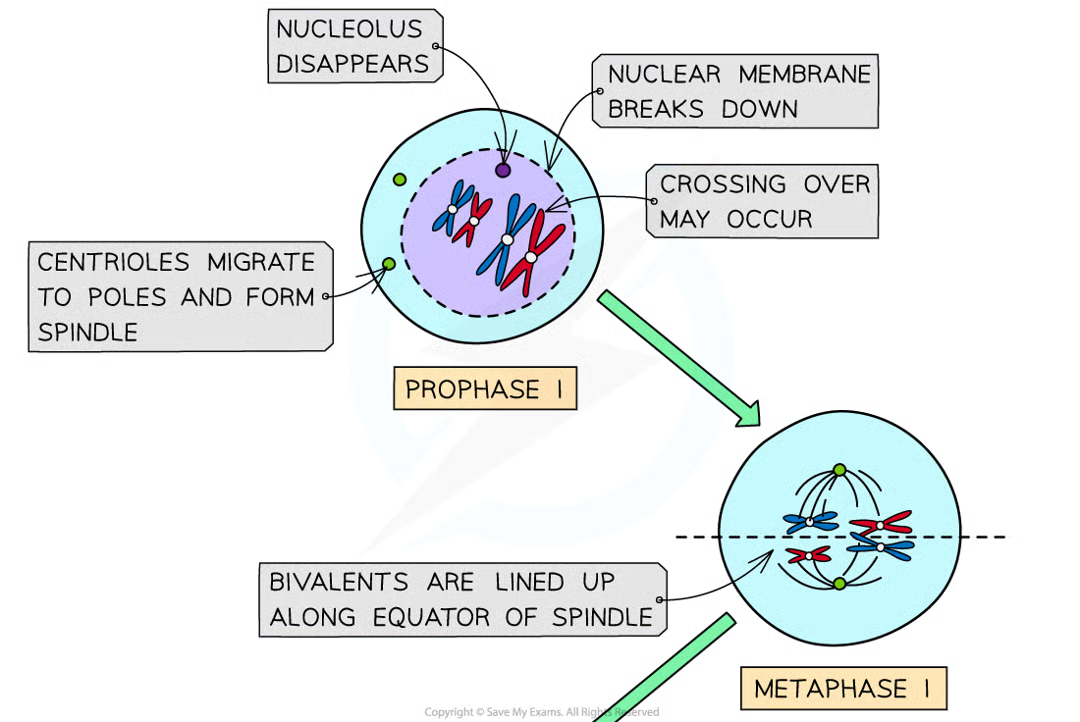
metaphase I 1
bivalents line up along equator of spindle, w spindle fibres attached to centromeres
anaphase I 2
homologous pairs of chromosomes are separated as microtubules pull whole chromosomes to opposite ends of spindle
centromeres don’t divide

telophase I 4
chromosomes arrive at opposite poles
spindle fibres start to break down
nuclear envelopes form around 2 groups of chromosomes and nucleoli reform
some plant cells go straight into meiosis II w/o reformation of nucleus in telophase I

cytokinesis overall, animal cells, plant cells, end product
when division of cytoplasm occurs
cell organelles get distrubted between 2 dev cells
animal:
csm pinches inwards creating cleavage furrow in middle of cell which contracts dividing cytoplasm in half
plant:
vesicles from golgi gather along equator of spindle (cell plate). vesicles merge w each other to form new csm. layers of cellulose are laid down to form primary and secondary walls of cell
end prod of cytokinese in meiosis I: 2 haploid cells which contain half the no. centromeres
what happens going into meiosis II
no interphase between I and II so dna not replicated
2nd division almost identical
p II 2
nuclear envelope breaks down and chromosomes condense
spindle forms at right angle to old one
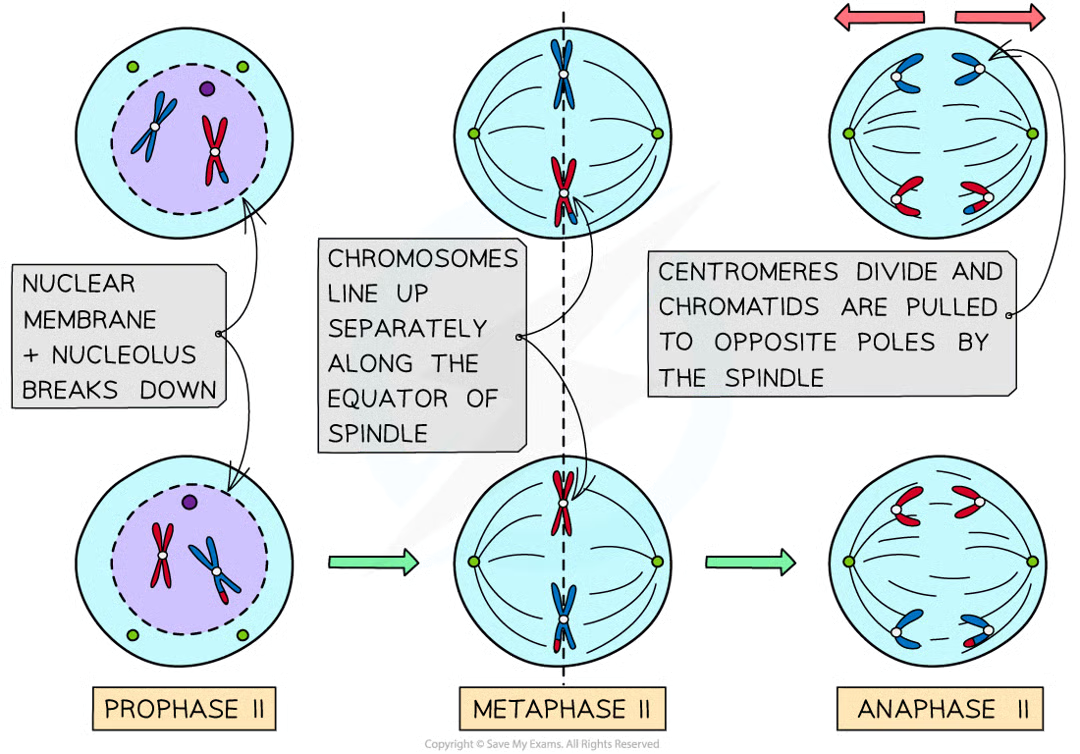
m II (1)
chromosomes line up in single file along equator of spindle
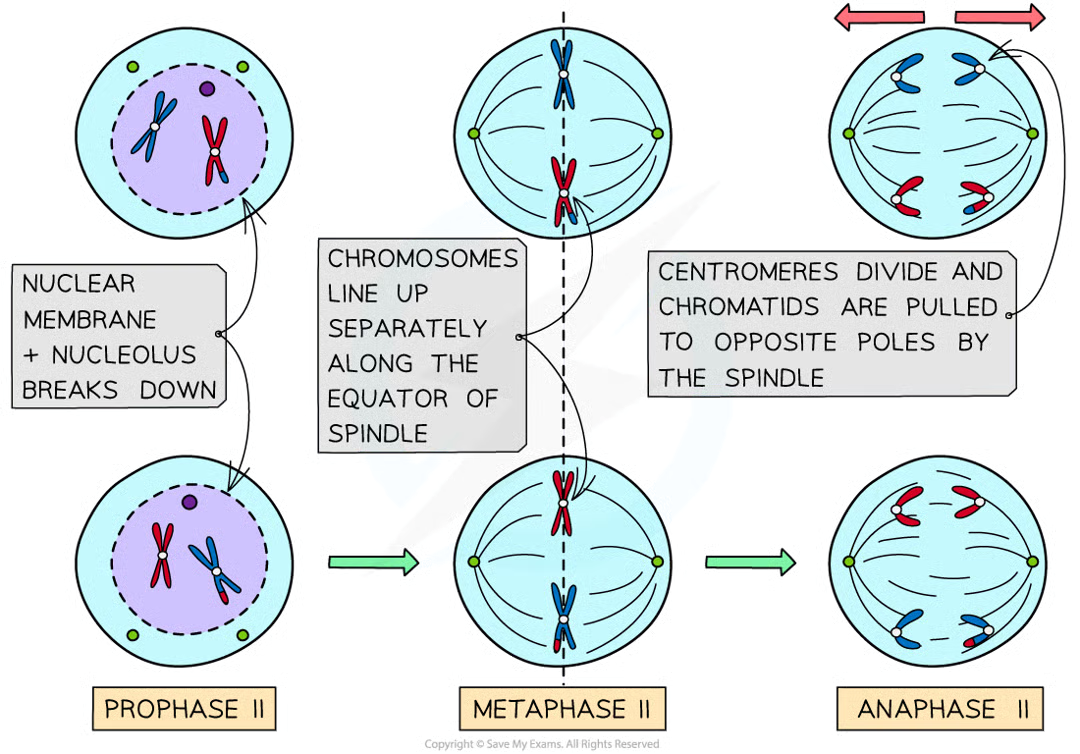
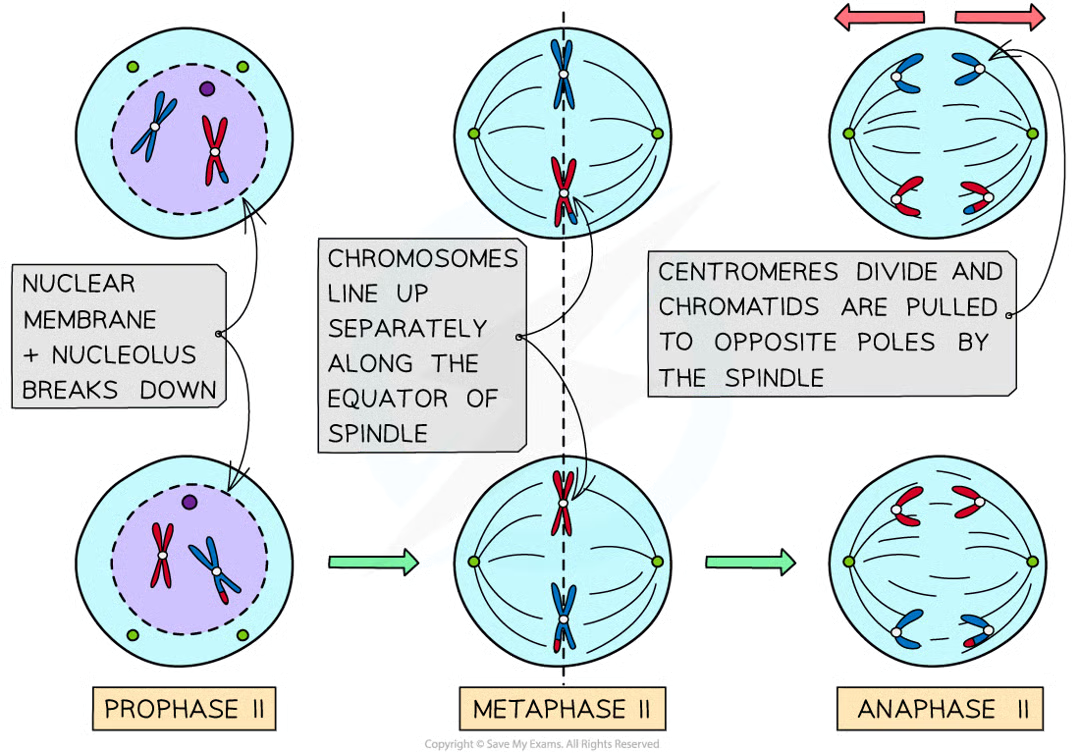
anaphase II (2)
centromeres divide and individual chromatids pulled to opposite poles
creates 4 grps of chromosomes that have half no. chromosomes compared to og parent cell
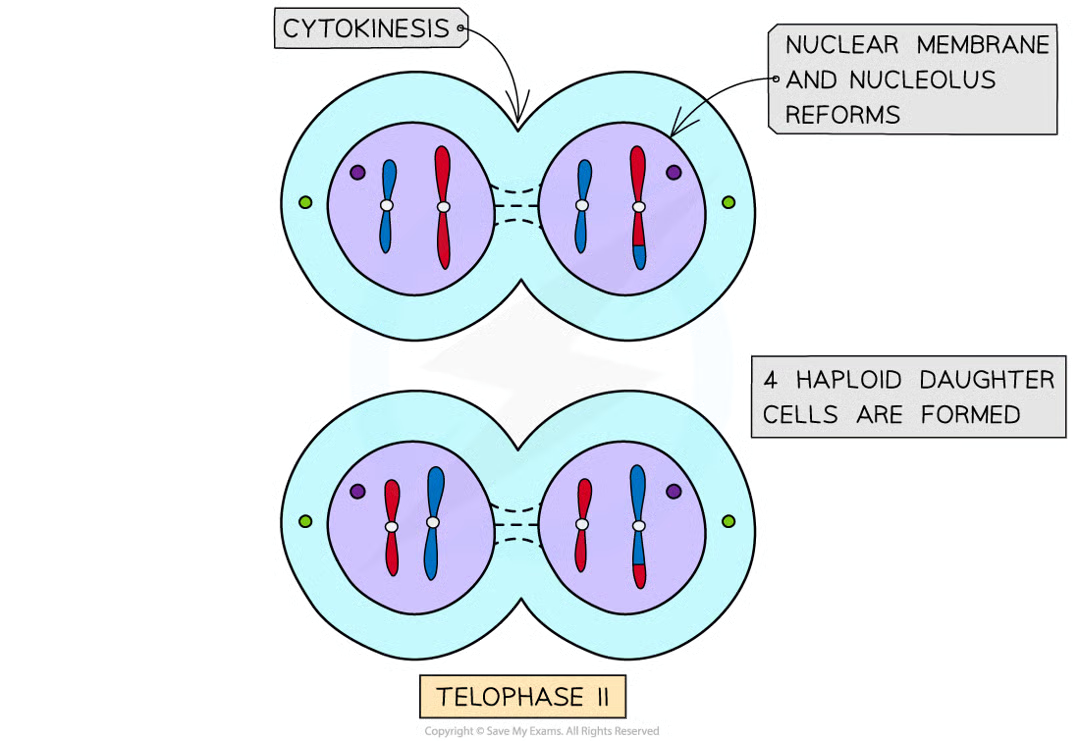
telophase II (1)
nuclear memb form around each grp of chromosomes
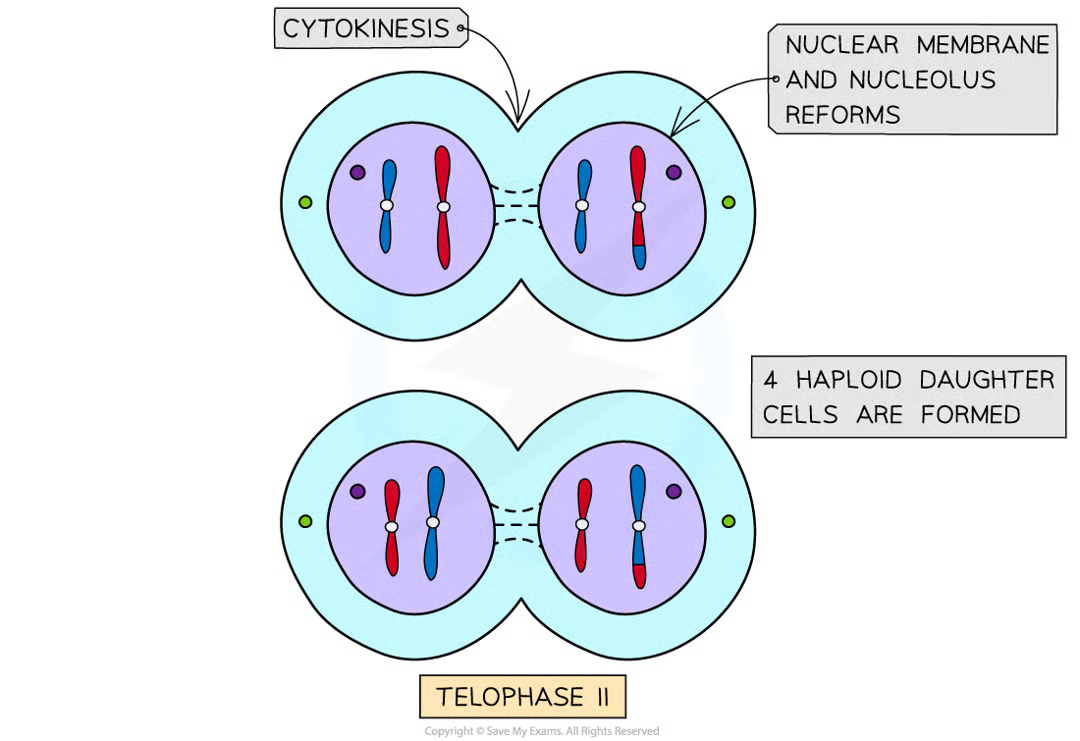
cytokinesis (after meio2)
cytoplasm divides as new csm formed creating 4 haploid ceclls.
cells still ctntain same no. centromeres as at start of meiosis I but now only half no. chromosomes (previoulsy chromatids)
how can stages of meiopsis be identified
cells can be observed and photographed using specialised microscopes
diff stages have distinctive characteristics meaning can be idenitifed from photomicrographs or diagrams
meiosis I vs II
homologous chromosomes pair up side by side in meiosis I only
if there are pairs of chromosomes in diagram/photomicrograph = meiosis I
no. cells forming can distinguish I and II
if 2 new cells forming = meio I but if 4 meio II
distinguishing features at each stage if meio I
pI: homologous pairs of chromosomes visible
mI: homologous pairs lined up side by side along equaotr of spindle
aI: whole chromosomes being pulled to opposite poles w centromeres intact
TI: 2 groups of condensed chromosomes around whcih nuclei membs forming
c: cytoplasm dividing and cellm memb pinching inwards forming 2 cells
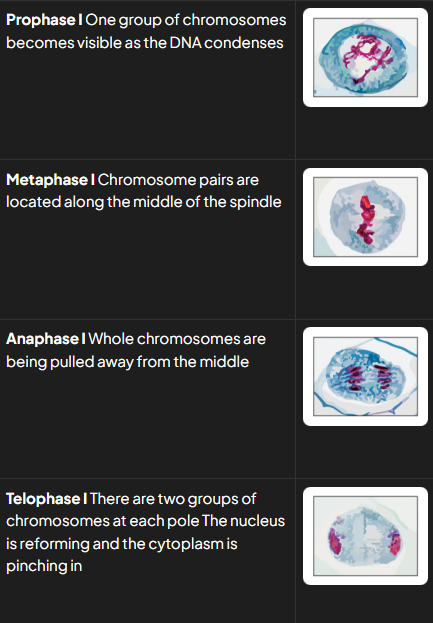
distinguishing features at stages of meio II
pII: single whole chromosomes visible
mII: single whole chromosomes lined up along equator of spindle in single file (at 90 degree angle to old spindle)
aII: centromeres divide and chromatids are being pulled to opposite poles
tII: nuclei forming around 4 groups of condensed chromosomes
c: cytoplasm dividing and 4 haploid cells forming
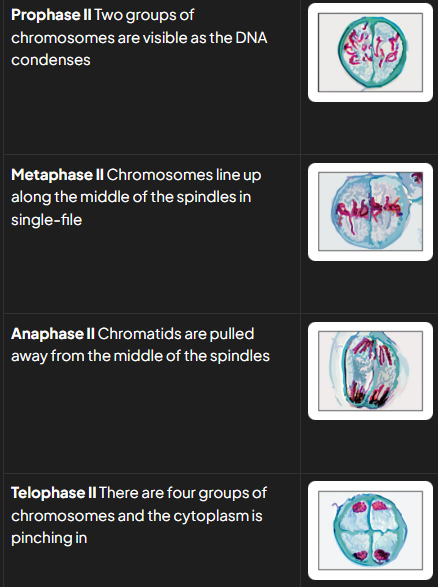
pmat tips
p: preparing to dviide
m: middle of spindle and cell where chromosomes line up
a: away from middle to poles where chromosomes pulled
t: two cells (for meiosis I at least)
why is genetically diff offspring advantageous
for natural selection
meiosis has several mechanisms that do what
increase genetic diversity of gametes produced. the following reuslt in diff combinations of alleles in gametes
crossing over
independent assortment
crossing over
process by which non-sister chromatids exchange alleles
process of crossing over
during meio I homologous chromosomes pair up and are very close in proximity to each other
non-sister chromatids can cross over and get entagled
crossing points = chiasmata
entaglement places stress on dna mols
as a result section of chromatid from one chromosomes may break and rejoin w chromatid from other chromosome
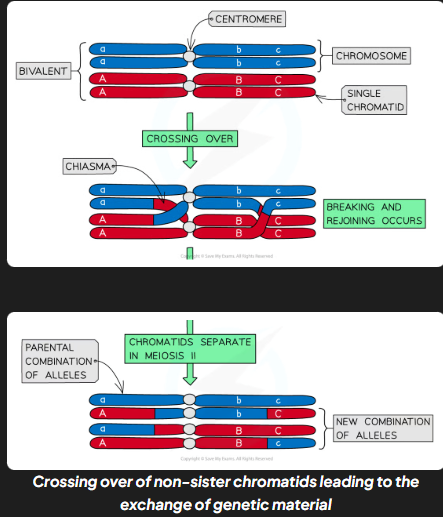
what does crossing over result in
swapping of alleles results in new combo of alleles on 2 chromosomes
usually at least one if ont more chiasmata present in each bivalent during meiosis
crossing over more likely to occur further down chromosome away from centromere
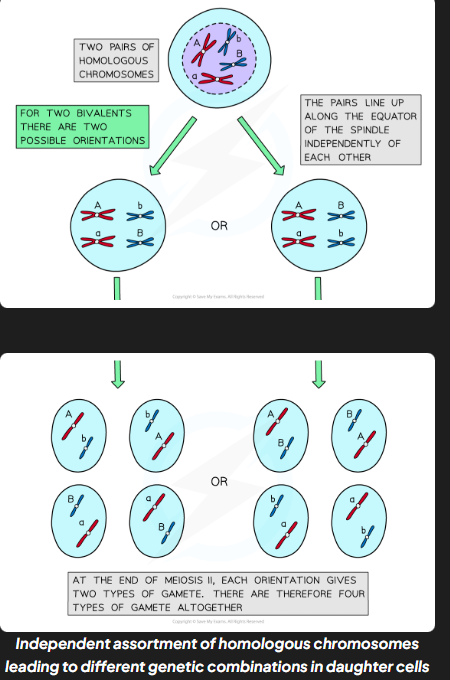
independent assortment
production of different combinations of alleles in daughter cells due to random alignment of homologous pairs along equator of spindle during mI/
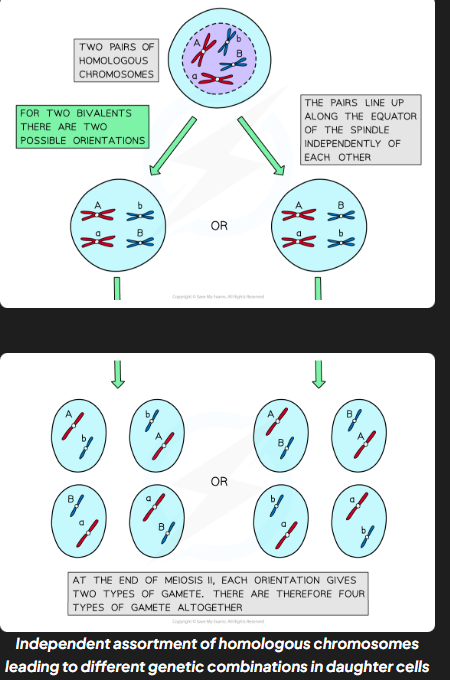
why does IA increase genetic variation
diff combinations of chromosomes in daughter cells increase genetic variation between gametes
IA process
in pI homologous chromosmoes pair up and in mI they are pulled towards the equator of the spindle
each pair can be arranged w either chromosome on top (completely random)
orientation of one homolgous pair is independent (unaffected by orientation of any other pair)
homologous chromosmes separated and pulled apart to diff poles
combination of alleles that end up in each daughter cell depends on how pairs of homologous chromosmes were lined up
to work out no. diff possible chromosomes combinations the formula 2n can be used where n corresponds to no. chromosomes in haploid cells (for huamns 2²3 - 8,388,608 diff combos.
fusion of gametes
meiosis creates gen variation between gametes produced by individual crossing over and independent assortment meaning each gameete has substantially diff alleles
during fert any male gamete can fuse w any female gamete to form zygote
random fusion of gametes at fert creates gen variation between zygotes as each will have a unique combination of alleles
almost 0 chance of individual organisms resulting from successive sexual reprod being genetically identical

DNA
dna contained within chromosomes essential for cell survival
every chromosome consists of long dna mol which codes for several diff proteins
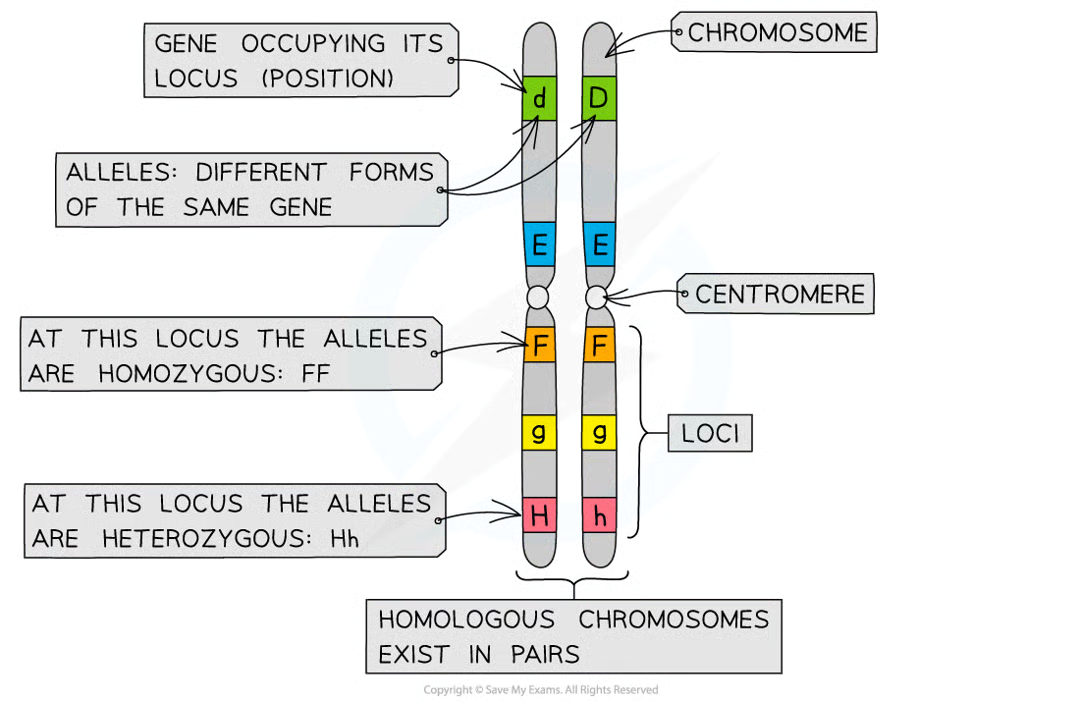
gene
length of dna that codes for single ppt or protein = gene
locus/loci
position of gene on chromosome
alleles
each gene can exist in 2+ diff forms = alleles. each have slightly diff nucleotide sequences but still occupy same position (locus) on chromsomes
example of alleles
one of genes for coat colour in horses is Agouti
gene found on same position on same chromosome for all horses
2 diff alleles of genes found in horses A and a
each allele can produce diff coat colour:
allele A → black coat
allele a → chestnut coat
genotype
refers to alleles of a gene possessed by that individual. the diff alleles can be represented by letters
why are there diff allele combinations within individual
chromosomes of eukaryotic cells occur in homologous pairs (2 copies of each chromosome)
as a result → two copies of every gene
as there are 2 copies of a gene present in individual = diff allele combos
homozygous
when 2 allele copies are identical in an individual
heterozygous
when 2 allel copies are diff in indiviudal
genotype affects….
phenotype
phenotype
observable characteristics of organism
example of genotype and phenotype
every horse has 2 copies of coat colour gene in all of cells
a horse that hsa two black coat alleles A has genotype AA and is homozygous so phenotype is black coat
horse w heterozygous Aa phenotype is black coat
horse with aa homozygous recessive = chestnut phenotype
do all alleles affect phenotype in same way
no, some are dominant CAPITAL, some are recessive lower case
dominant:
always expressed in phenotype meaning expressed in both heterozygous and homozygous
recessive:
only expressed if no dominant allele present
only w homozygous
example of donminance
if A for black and a for chestnut following:
AA → black coat
Aa → black coat
aa → chestnut
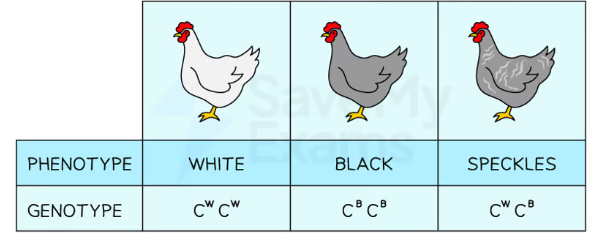
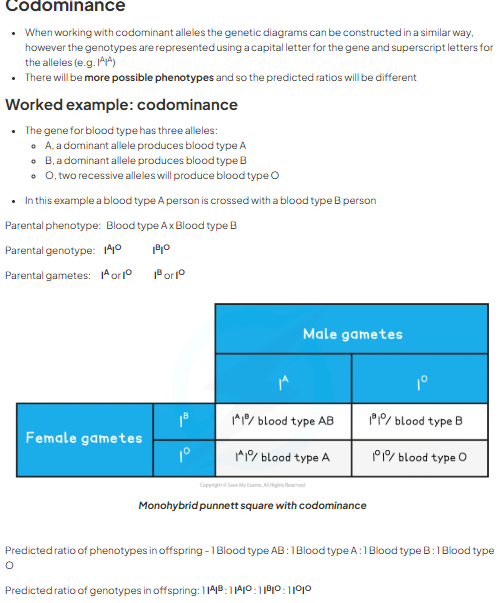
codominance examples
blood group
gene for blood types represented by I and three alleles = A, B, O. allele for O is recessive to A and B. A results in A (IAIA or IAIO) and B results in (IBIB or IBIO). if both a and b present in heterozygous = AB IAIB
feather colour in picture
f1 generation
when homozygous dominant individula crossed w homozygous recessive individual, offspring = f1 generation, all heterozygous
f2 generation
if 2 individuals from f1 gen are then crossed, the offspring produce f2 generation
test cross
can be sued to try and deduce genotype of unknown individual that is expressing dominant phenotype
indiviudla in question is crossed w indivudla expressing recessive phenotype
resulting phenotypes of offpsring provide sufficient info to suggest genotype of unknown
if any offspring expressive recessive then unkown must be heterozygous
2 types of linkage
sex linkage and autosomal linkage
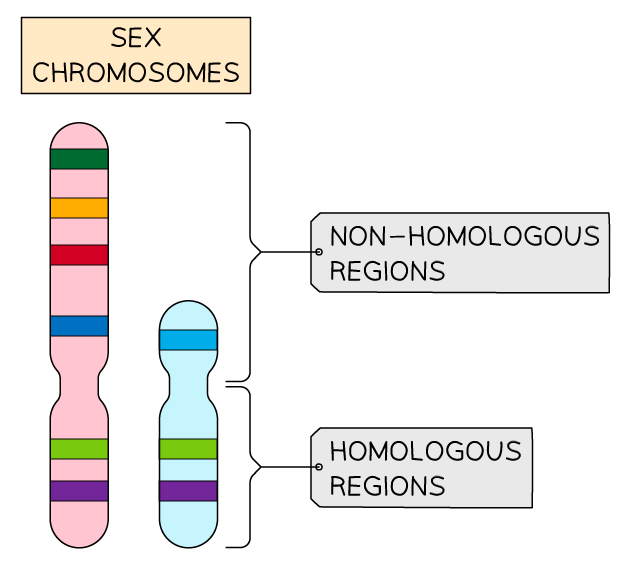
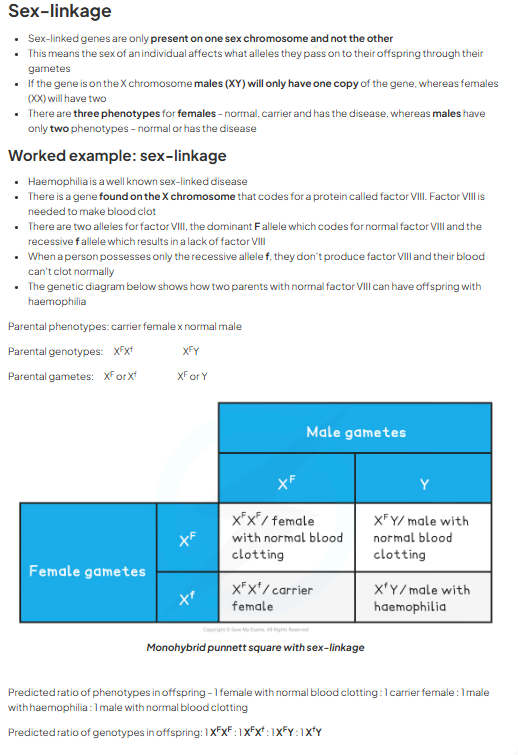
sex linkage:
two sex chromosomes X and Y
women (XX) men (XY)
some genes found on region of sex chromsome X that is not present on sex chromosome Y = non homologous region
as inheritance of genes dependent on sex of individual = sex linked genes
most found on longer X chromosome e.g. haemophilia
represented in genotype by writing alleles as superscript next to sex chromosmes (XGXg)
autosomal linkage:
occurs on autosomes (chromosome not sex chromosome)
two or more genes on same chromosome do not assort independently during meiosis
genes linked and stay together in og parental combo
monohybrid inheritance
looks at how alleles for single gene are passed on from one generation to the next
known info abt geno, pheno and process of meio used to predict pheno from specific breeding pairs
when 2 individuals sexually reproduce, theres equal chance of either allele from homologous pair making it into their gametes and subsequently the nucleus of zygote
equal chance of zygote inheriting either allele frrom parent
genetic diagrams
often used to present this info in a clear and precise manner so that predictions can be made
punnett square
are predictions accurate
no, based on chance, no way to predict which gametes will fuse so sometimes observed or real life results can differ
genetic diagram worked example

codominance worked example
sex linkage worked example
dihybrid crosses
look at how the alleles of 2 genes transfer across generations. similar to monohybrid but more phenotypes nad genotypes. must not mix alleles from diff genes
worked example: dihibiryd genetic diagram
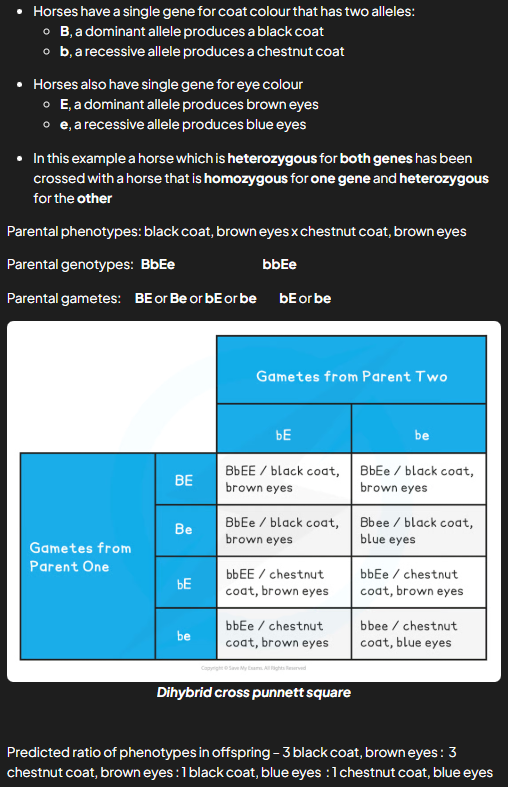
what do dihibryd cross predictions rely on
assumption that genes being investigated behave independently of one another during meiosis
do all genes assort independently
no
what do some genes display
autosomal linkage where genes are located on same chromosome and stay together in the original parental combination which affects how parental alleles are passed to offspring through gametes
how to display autosomal linkage when writing
(FG) (FG) put in brackets instead of FFGG
worked example autosomal linkage

epistasis
when two genees on different chromosomes affect the same feature. dictates phenotype
epistasis worked example

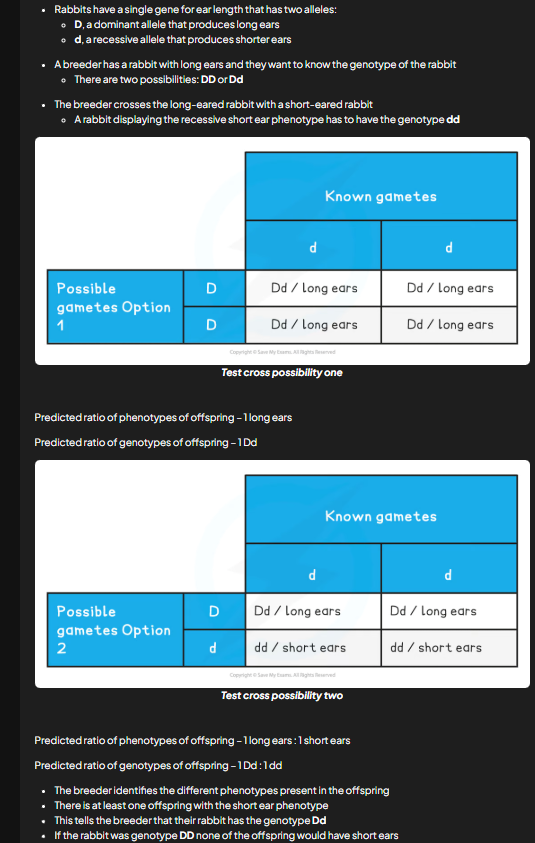
test cross
can be used to deduce the genotype of an unknown individual that is expressing a dominant phenotype.
how is test cross carried out
individual is crossed w individual expressing the recessive phenotype because it has a known genotype and resulting phenotypes provide sufficient info to suggest unknown gtype
result of monohybrid test cross (2)
if no offspring exhibit receessive phenotype then unknown is homozygous dominant
if at least 1 of the offspring exhibit the recessive phenotype then the unknown genotype is heterozygous
result of dibhybrid test cross (3)
if no offpsring exhibit recessive phenotype for either gene then the unkown genotype is homozygous dominant for both genes
if at least one of the offspring exhibit the recessive phenotype for one gene but not the other then the unkonw genotype is heterozygous for one gene and homozygous dominant for another
if at least one of the offspring exhibit recessive phenotype for both genes then unknown genotype is heterozygous for both genes
test crosses worked example
why can there be a difference between expected and observed results in experiments
can be statistically significant or insignificant (happened by change). if the difference between results is statistically significant it can suggest that something else is happening in the experiment unaccounted for e.g. linkage
what does chi-squared test determine
whether there is a significant difference between the observed and expected results in anexperiment
when is a chi squared test completed
when data is categorical (data that can be grouped)
how to calculate chi-squared values
obtain the expected (E) and observed (O) results for the experiment
calculate the difference between each set of results
square each difference (+ or - irrelevant)
divide each squared diff by the expected value
add the resulting values together to get a sum of these answers to obtain the chi-squared value
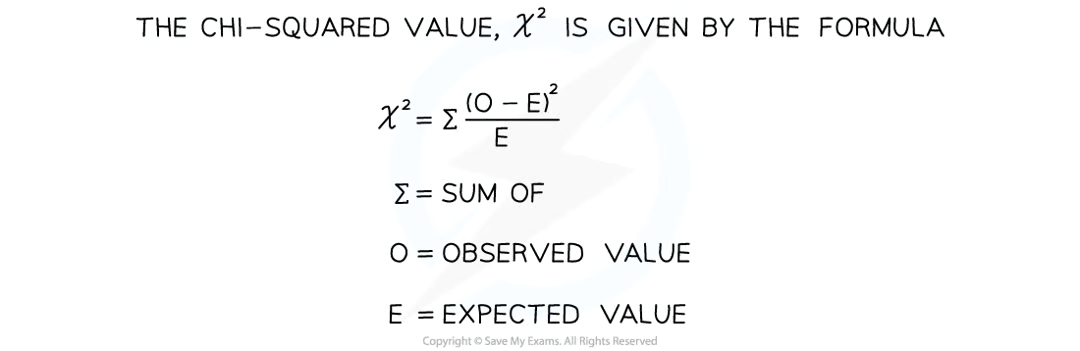
how can we work out what chi squared value means
we need to compare the chi-squared value to a critical value
where is the critical value read from
a table of critical values and depends on the probability level used and the degrees of freedom
what probability level do biologist use
0.05/5% probability that dif fbetween O&E is chance
how is degrees of freedom calculated
number of classes - 1 e/g/ 2 phenotypes = 2-1=1 degree of freedom
if chi squared greater/equal to CV then..
there is significant difference between O and E so a factor other than chance is causing the difference meaning a null hypothesis can be rejected.
if chi squared value is smaller than CV then…
no sig diff between o and e, diff due to chance and null hypothesis accepted
e.g. of using cv table ot assess probability at which any diff between o and e significant
chi squared value might be smaller than cv at prob of 0.05 but larger at 0.1 so chance prob between 5-10%.
chi squared might be larger than crit value at prob level of 0.001 indicating that less than 0.1% probability that any diff between o and e is chance
what can gene affect and how
phenotype as it codes for a single protein that affects phenotype through particular mechanism (environment also affects it)
albinism
when humans lack pigment melanin in skin hair and eyes causing them to have very pale skin, very pale hair and pale blue or pink irises in the eyes
metabolic pathway for producing melanin
aa tyrosine is converted to DOPA by enzyme tyrosinase
DOPA converted to dopaquinone again by enz tyrosinase
dopaquinone converted to melanin
tyrosine → DOPA → dopaquinone → melanin
what happens to TYR gene with albinism
TYR gene on chromosome 11 codes for enzyme tyrosinase
recessive allele for TYR causes lack of enz tyrosinase for presence of inactive tyrosinase
without tyrosinase enz tyrosine cant be converted into melanin
sickle cell anaemia
condition that causes idnividuals to have frequent infections, episodes of pain and anaemia due to abnormal haemoglobin in RBC
why does sickle cell anaemia happen in the body
beta-globin is a ppt found in haemoglobin that is coded for by the gene HBB which is foudn on chromosome 11.
there’s an abnormal allele for the gene HBB which produces a slightly different aa sequence to the normal allele
the change of a single base in the DNA of the abnormal allele results in aa substitution
dna base sequence GAG is replaced by GTG and CTC is replaced by CAC on the complementary DNA template strand so GAG is replaced by GUG in the resulting mRNA
this results in abnormal beta-globin ppt as aa Glu is replaced with Val
what happens to rbc in sickle cell
the abnormal beta globin in haemoglobina ffect the structure and shape of the RBC
pulled into half moon shape
unable to transport oxygen around body
they stick to each other and clump together blocking capillaries
what happens if a homozygous individual that has two abnormal alleles for the HBB gene
produces only sickle cell haemoglobin and will have sickle cell anaemia and suffer from the associated symptoms
what happens if a heterozygous individual that has one normal allele and on abnormal allele for the HBB gene
will produce some normal haemoglobin and some sickle cell haemoglobin
they are carrier of allele
they may have no symptoms
