Protein shape
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
46 Terms
primary structure
linear sequence of amino acids residues
determined by mRNA code
in combination with protein’s environment determines secondary, tertiary, quaternary
is the amino acid sequence of every protein identical to the genetically encoded primary sequence?
yes
secondary structure
folding and twisting of peptide backbone
held together by weak H-bonds between C=O and N-H groups in backbone
R-groups stick out from backbone
two well-known secondary structures: alpha helices and beta sheets
alpha helix
rigid cylindrical structure
forms when H-bonding occurs between a C=O and N-H groups that are 4 amino acids apart on polypeptide backbone
coiling happens in a clockwise direction down the length of chain
beta sheet
flat sheet like structure
form when H bonding occurs between C=O and N-H groups on adjacent polypeptide chains
what do rigid proline residues do for protein backbone?
it inserts a kink in and disrupts secondary structures
tertiary structure; its held together by?
3D arrangement of secondary structures
mostly held together by noncovalent attractions between:
R-groups
between R-groups and the surrounding environment (ie aqueous or hydrophobic lipid bilayer interior)
unstructured loops (aka random coils)
link secondary structures together
what do covalent disulfide bonds do for cysteine residues
they cross-link parts of the polypeptide backbone
3D folding of proteins into results in structures that assume the —
lowest possible energy state
protein stability depends on —
the free energy change between the folded and unfolded states; deltaG = Gfolded - Gunfolded
proteins become more stable as Gunfolded — Gfolded
>
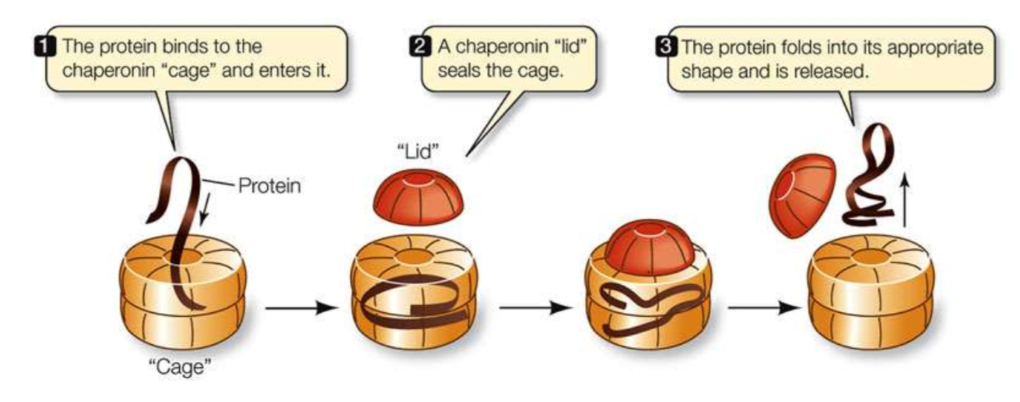
chaperonins
molecular chaperones that many proteins require that provide an isolated chemical environment in which they can fold
steps of chaperonin bondings
prions
a disease of protein folding
prion proteins can adopt an alternative folded state
abnormally folded protein causes a normally folded protein to adopt the abnormal conformation
protein domains
a region of the protein that folds independently of other regions
protein can have single/multiple domains
domain often represents a functional region of the protein
function of catalytic domain
inhibits host cell protein synthesis
function of receptor binding domain
attaches to cell surface
function of hydrophobic domain
inserts into membranes
motifs
similar domain which occur in many related proteins ex. DNA-binding motif
quaternary structure
arrangement of multiple tertiary structures
held together by weak bonds and some disulphide bonds
homomers
identical subunit polypeptidesd
heteromers
different subunit polypeptides
proteolytic cleavage
removes amino acids from the original translated sequence
protein kinases
a class of enzymes that catalyzes reactions
protein phosphatases
catalyzes phosphate removal; reverses phosphorylation
many changes in protein structure and activity are driven by —
phosphorylation
each phosphate group adds — to the protein
two negative charges
the added phosphate group may create a — — — that allows other proteins to bind to the phosphorylated protein
new recognition site
many changes in protein structure and activity are driven by phosphorylation because:
each phosphate group adds two negative charges to the protein
Can drive major structural changes, activity changes, or changes in protein solubility
Added phosphate group may create a new recognition site that allows other proteins to bind to the phosphorylated protein
You are studying a protein that is turned on by phosphorylation at a specific serine residue. What do you expect would happen if the serine is mutated to aspartic acid?
the protein will always be turned on
ubiquitin
small cytosolic protein (76 amino acids)
covalently attached to proteins (reversible)
serves as tag that can either mark proteins for degradation or direct proteins to specific locations in the cell
(strand: degradation; singular: translocation)
all proteins bind to other molecules, those molecules are called - of the protein in question
ligands
a protein’s physical interaction with other molecules determines —
its biological properties
because ligand binding is generally achieved by noncovalent bonds it is —
reversible
ligand binding is generally achieved by — bonds
noncovalent
why must protein binding be strong enough to withstand the jolting of molecular motions
molecules are in constant motion, bumping into one another
binding strength is achieved through
3D complementarity of binding
formation of several noncovalent bonds (strength in numbers)
ligand binding sites are — dimensional
3
amino acids that contribute to binding a liganf are often far apart on — but come together when —
a proteins primary sequence; protein folds
lower dissociation rates =
lower Kd values and stronger binding
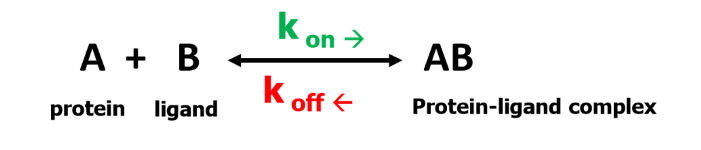
Kon and Koff are rates of the — and — rxns that create/breakdown protein ligand complex
forward (association); backward (dissociation)
association Ka measures the strength of binding such that
Ka = Kon/Koff
relationship of dissociation constant to association constant
Kd=1/Ka
having multiple modification/interaction sites allows proteins to act as —
molecular integrators