VIRO 3 | Models for Virus Replication
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
16 Terms
Explain general viral replication strategies
All viruses need to hijack host protein synthesis machinery to produce viral proteins and new virions
But replication strategies differ depending on type of genomes they carry: DNA, RNA, RT DNA/RNA
dsDNA = Class I
ssDNA = Class II
dsRNA = Class III
+ssRNA = Class IV
-ssRNA = Class V
dsDNA RT = Class VI
ssRNA RT = Class VII
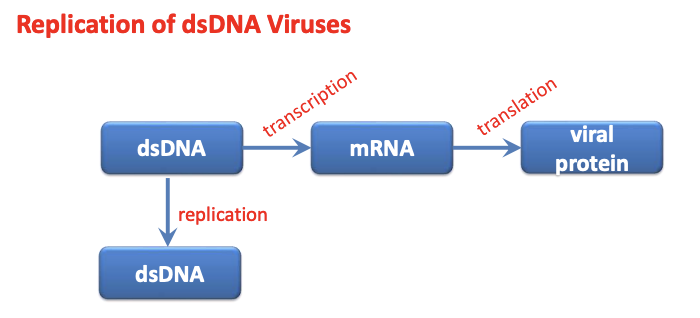
Explain general DNA virus replication strategies, Herpesvirus as dsDNA virus (Class I)
General DNA virus replication strategies
Virus needs to transcribe mRNA, which can the be translated into proteins via host cell translation machinery
dsDNA > mRNA > viral protein
Virus needs to replicate its own genome
dsDNA > dsDNA
Some DNA virus can replicate in cytoplasm using viral DNA polymerase
For those who can’t, host enzymes for mRNA synthesis & DNA replication are in nucleus; to avail of these enzymes, they must enter nucleus
Herpesvirus = dsDNA virus
Viral structure
Enveloped virus with linear dsDNA genome
Multiple protein layers ntv
At its core = nucleocapsid containing linear dsDNA genome
Protein tegument layer = viral proteins with regulatory functions
Viral envelope glycoproteins = can act as TFs for immediate early gene expression
Genome features
Long
Terminal repeat sequences (L,S)
Unique sequences are separated by inverted repeat sequences (L,S)
Entry & transport to nucleus
Once HSV binds to its receptor, viral fusion protein facilitates fusion of viral envelop + plasma membrane, allowing nucleocapsid + tegument proteins to enter
Nucleocapsid then uses host cytoskeletal elements (microtubules & motor proteins dynein & kinesin) to reach nucleus
Once at nucleopore, it inserts genome into nucleus, where it is circularized to prevent end replication problem & ensure complete genome replication
Transcription initiation & immediate early gene expression
Some tegument proteins enter nucleus via NLS
These act as transcription factors for immediate early genes, recruiting host RNA pol II to transcribe viral mRNA
Early gene expression & DNA replication
Immediate early genes trigger expression of early genes, including gene for viral DdDp
mRNA for DdDp is exported to cytoplasm for protein translation
then imported back to nucleus (via NLS), where it will then replicate viral genome
Virus replicates via rolling circle replication strategy, producing concatemers = long DNA strands with multiple genome copies linked together
This is later on cut and packaged into capsids
Capsid assembly & genome packaging
Capsid & scaffolding proteins help in forming maturing nucleocapsids
Viral genome is packaged into nucleocapsids
Packaging signal in viral genome specify which length/segment is included
Assembly > nucleus, structural proteins must be imported into nucleus
Envelopment & maturation
Assembled nucleocapsid first buds into nuclear membrane, acquiring temporary envelop
It enters ER, removing & re-acquiring envelop
Nucleocapsid buds into a vesicle, carrying envelop + viral glycoproteins
Vesicles moves into Golgi for further processing
Golgi = where it acquires viral glycoproteins
Virion is transported into host cell membrane; vesicle fuses with HCM, releasing mature enveloped virion out of host cell via exocytosis
CPE = Syncytium formation
HSV causes syncytium formation (glycoproteins on surface of infected cells bind to neighboring cells > fusion) as CPE
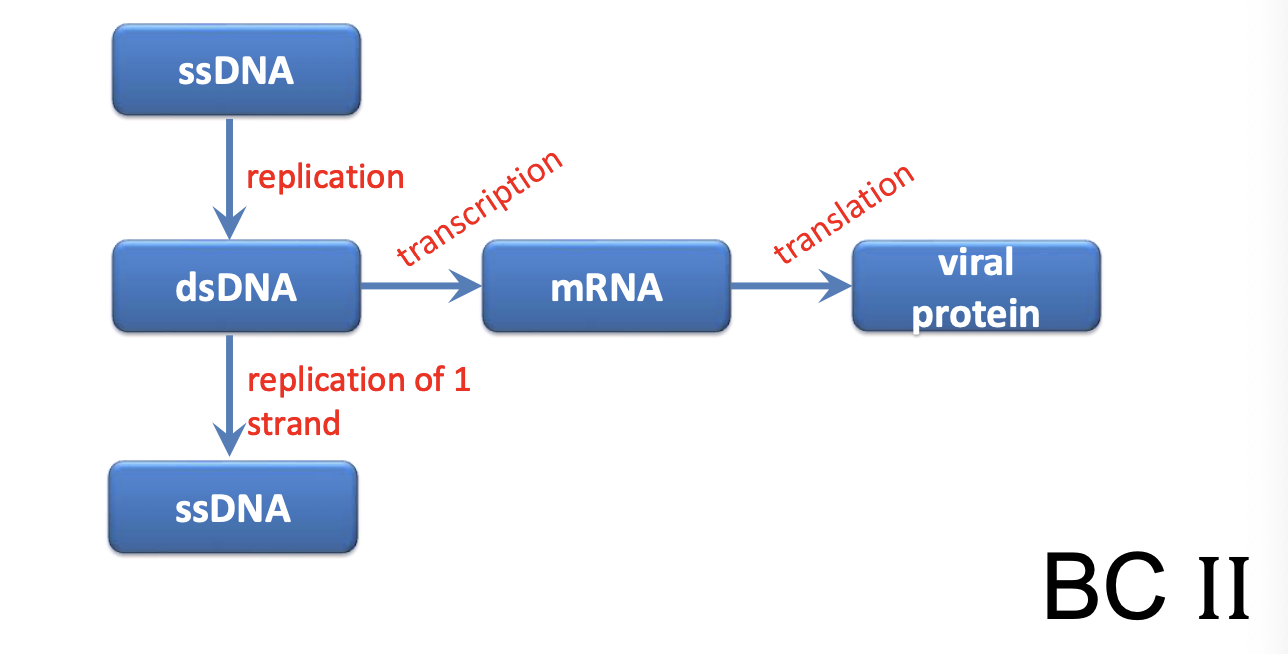
Explain Parvovirus as ssDNA virus (Class II)
General properties
“Parvo” = small; smallest known DNA viruses
Nucleocapsid + ssDNA genome
Genome = 9 viral genes encode 9 viral proteins (both structural & nonstructural)
Bc of its small genome, it relies heavily on host cell machinery
Tropism
It targets rapidly dividing cells bc it needs active host DNA polymerase abundant during S phase of cell cycle
In dogs, they infect gut epithelial cells
In humans = precursors of blood cells (in bone marrow)
Dependency: AAV & co-infection
Adeno-associated virus (AAV) = kind of parvovirus that requires co-infection w/ adenovirus to replicate
AAV = not free-living
Not pathogenic and thus widely used in gene therapy, used to deliver foreign genes safely into human cells
Entry & transport into nucleus
Upon binding to receptors, Parvo is internalized via receptor-mediated endocytosis
Has phospholipase and thus degrades endosomal membrane, escaping into cytoplasm
Uses dynein & kinesin to reach nucleus
Bc of its small size, entire nucleocapsid enters nucleus
Genome structure & rolling hairpin replication
Genome is flanked by ITR that forms hairpin structure (forms 3’OH) crucial for replication, as it is recognized by DNA polymerase to initiate synthesis of complementary DNA strand
Genome is not circularized and thus not directly used by host polymerases
Virus does “rolling hairpin” replication strategy, where ITRs provide 3’OH recognized by DNA polymerase to initiate synthesis of complementary DNA strand
1 strand acts as template to generate more genomes
Both viral replication & transcription occurs in nucleus
Gene expression & protein handling
Viral mRNA is exported into cytoplasm for protein translation
Nonstructural proteins > nickase cuts 1 strand of DNA to expose 3’ OH region for DNA synthesis
Structural proteins (capsid proteins) are then imported back to nucleus for several reasons:
Since viral genome replication occurs in nucleus, virion assembly = nucleus
Structural proteins risk degradation when left in cytoplasm
Cells treat DNA as threat in cytoplasm; degraded by nuclease
Importing proteins back to nucleus helps protect DNA & support virion assembly
Virion assembly & exit
Once virions are assembled, these are released via lysis, bursting host cell & releasing viral particles
Explain overview of RNA virus replication strategies
Classes
IV (dsRNA) = Rotaviruses, Reoviruses
dsRNA cannot function as mRNA
require RdRp in virion
bc dsRNA is PAMP (pathogen-associated molecular pattern; molecular structure broadly shared among pathogens but not normally found in host cells and thus would trigger immune response) and thus mRNA would need to be made within protected environment
V (+ssRNA) = COVID-19, dengue
+ssRNA can function as mRNA and thus would not need RdRp in virion
Rather, +ssRNA can act directly as mRNA and encode RdRp later for use
VI (-ssRNA) = Influenza, rabies
-ssRNA cannot function as mRNA
require RdRp in virion
RdRp is needed to make mRNA from negative strand
RNA viruses do not have a DNA phase, unlike retroviruses
RNA viruses bypass normal cell processes bc cells don’t usually use RNA as template to make more RNAs
Instead, they rely on RNA-dependent RNA polymerase, which is a viral protein that doesn’t have a cellular equivalent; only virus can encode it
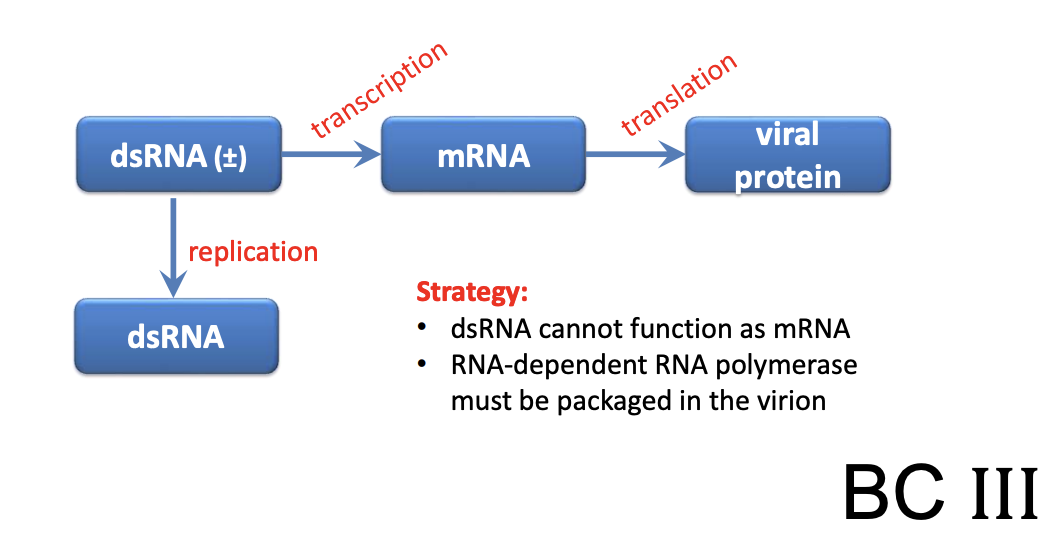
Explain Rotavirus as dsRNA virus (Class III)
Structure
Has dsRNA genome = PAMP and thus would trigger immune responses > TLR
Has multilayered capsid = inner, intermediate, outer protein layer to keep dsRNA hidden from host cytoplasm
Entry & uncoating
Upon binding to receptors, Rotavirus is internalized via receptor-mediated endocytosis
Within endosome, outer protein layer is degraded, thus releasing a double-layered particle (DLP) into host cytoplasm
Transcription & replication
Transcription begins in DLP to hide dsRNA from cytoplasm
Each RNA segment is associated with RdRp; mRNA is transcribed then extruded into cytoplasm for protein translation
+RNA transcript = template for protein synthesis; dsRNA replication happens within protected particle
Assembly & maturation
Newly synthesized viral proteins & RNA segments are assembled into DLPs in viroplasm
dsRNA synthesis occurs in assembling DLPs
DLPs acquire another protein coat as it passes thru rough ER, which will be proteolytically cleaved as they exit host cell upon maturation → triple-layered particle (TLP)
Design ensures dsRNA = never exposed to host cytoplasm
Host defenses = RNA interference (RNAi)
Host combat viral infection via RNAi
RNAi = post-transcriptional gene silencing mechanism that’s highly sequence-specific
Uses Dicer protein that cleaves viral dsRNA into shorter segments, then loads it into RISC
In the RISC,
One strand is degraded; the other is used as guide to bind to complementary mRNA
Perfect complementarity = mRNA being cleaved & degraded
Partial complementarity = translation is repressed; ribosomes cannot bind
RNAi = a mechanism that prevents viral protein production & antiviral mechanism across many organisms
RNA silencing therapeutic use
RNAi can be harnessed to target viral mRNA, causing either:
Post-transcriptional gene silencing
For COVID-19, scientists are introducing small interfering RNA (siRNA) that guide RISC to complementary viral mRNA, allowing it to be cleaved & degraded
Transcriptional gene silencing
For HIV, scientists are introducing synthetic dsRNA that activate histone deacetylases, causing DNA to tighten and not be read; thus mRNA is not produced
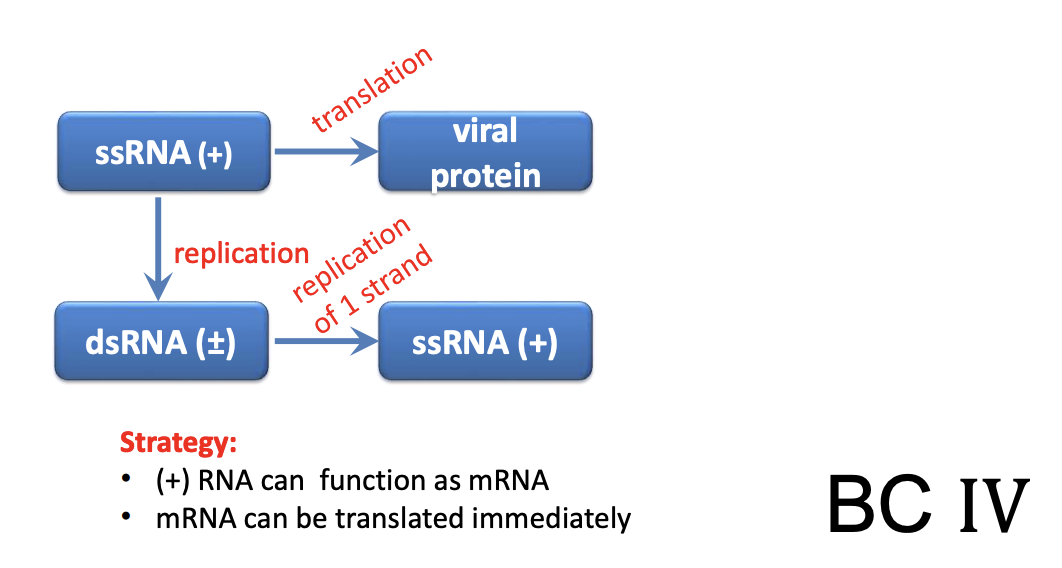
Explain +ssRNA (Class IV) viruses
General properties
+ssRNA can immediately act as mRNA
Upon entry into cell host, ssRNA can serve as mRNA & be immediately translated by host ribosome into
Polyproteins 1a & 1b > cleaved into nonstructural proteins > RdRp
+ssRNA thus don’t need RdRp packaged in virion
Instead, they encode it into their genome, such that after translation > RdRp is synthesized
Once RdRp is synthesized, viral genome replication occurs
RdRp uses +ssRNA as template to create -ssRNA
-ssRNA is used as template to create more -ssRNA genomes
-ssRNA can be converted into transient dsRNA
Complementary - strand of dsRNA is used as template to produce more +ssRNA for genome amplification to ensure there’s enough viral RNA for virion packaging
+ssRNA has multiple roles in virus life cycle
act as mRNA for immediate translation
template for synthesis of -ssRNA
template for synthesis of more +ssRNA for new virions
e.g., Picornavirus
“Pico” = small, RNA virus
Small, non-enveloped +ssRNA viruses
e.g., Poliovirus, rhinovirus
+ssRNA > mRNA transcript = no need for prepackaged enzymes
e.g., SARS-CoV-2
enveloped +ssRNA virus with large single genome
Genome encodes sps
Structural proteins: spike, envelope, membrane, nucleocapsid
Polyproteins (1a, 1ab): cleaved into NS proteins
Subgenomic RNA (copies of viral genome; accessory & structural protein-coding region) for expression of structural & accessory proteins
Significance of subgenomic RNA = allows for separation of early non-structural (NS) protein production then later structural (S) protein synthesis
mRNA translation produces polyproteins 1a & 1ab, cleaved into nonstructural protein > RdRp used for genome replication
6 nested subgenomic mRNAs with 5’ leader sequence produce structural proteins after replication & transcription begins
Fusion & Entry
Fusion between viral envelope & host cell membrane occur in 2 ways:
At cell surface, when proteases like TMPRSS2 activates spike protein
Within endosome, after endocytosis & spike activation in acidic vesicles
Once inside cytoplasm, translation begins immediately bc +ssRNA > mRNA transcript
Replication in DMV
Genome replication occurs in double-membrane vesicles, which has 2 critical functions
Immune evasion = DMV prevents host immune system from recognizing dsRNA intermediates
Efficient genome production = DMV concentrates replication machinery & templates within confined space, speeding up & increasing amount of viral mRNA synthesis for assembly
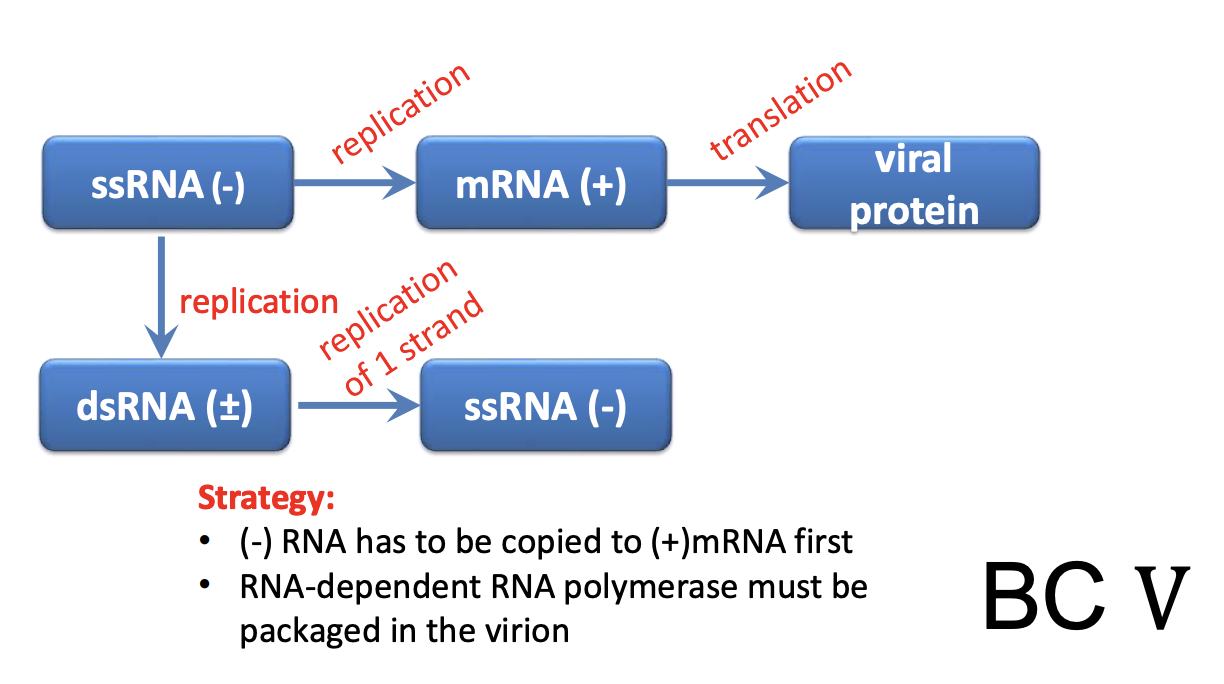
Explain -ssRNA viruses (Class V); Influenza viruses
Before the COVID-19 pandemic, Human Parainfluenza Virus (HPIV) was predicted to cause next outbreak
Influenza virus genome structure
IAV consists of segmented genome
8 RNA segments, from longest to shortest
3P H NNMN
PB2
PB1
PA
Hemagglutinin
Nucleoprotein
Neuraminidase
Matrix protein
Nonstructural protein
Each RNA segment is
packaged in nucleocapsid
capped w/ polymerase complex that includes endonucleases that snatches 5’ cap of host mRNA to prime mRNA synthesis
Each RNA segment encodes specific protein
Hemagglutinin = 18 subtypes & binds to sialic acid receptors on host cells
SA receptors are host-specific, affecting tropism
Important epidemiologically due to their role in host infection & immune recognition
Influenza pandemics & immunity
Multiple influenza pandemics have occurred, starting with 1918 flu
Caused by lack of natural immunity to new HN subtypes
Even slight changes in subtypes > immune evasion & outbreaks
Segmented genome & infectivity
Infectivity requires all RNA segments to be present
Packaging signals ensure correct assembly, enabling RNA-RNA & RNA-protein interactions
RNA-structural proteins
RNA-capsid proteins
RNA-envelop proteins
These interactions are important bc during budding, these ensure that all segments are included
Nucleocapsid are assembled perpendicular to host plasma membrane to ensure
Proper genome packaging
Correct interactions with envelope proteins
Efficient budding
Why segmented genomes matter > Reassortment
Bc segmented genomes can lead to reassortment that allow rapid evolution, particularly
If host is infected by 2 influenza viruses, the genome segments (H1N1, H5N1) can mix and result in strain that can evade immune recognition & cause outbreaks
Pigs are critical “mixing vessels”
Can be infected by both avian & influenza virus, thus reassortment may result in new strain that is transmissible to humans & can evade immune recognition
Antigenic shift vs. drift
Shift
Occurs when 2 strains infect same host cell, leading to reassortment & creation of new virions that can evade immune system > outbreaks
Drift
Involves random mutations over time due to error-prone RdRp that lacks proofreading abilities
Occurs in both segmented, non-segmented RNA genomes
Reduce overall antibody effectiveness
Multihost ecology of H5N1 (highly pathogenic influenza virus)
Reservoir host: aquatic birds; virus circulates stably among them
Spread to: poultry, seabirds, mammals, humans (occasionally)
Recently detected in: mink, cows, sea mammals, goat, cats
Global spread
Australia = only avian virus-free continent
Recent H5N1 developments in US
Cattle outbreak (2013)
Spillover from wildlife to cattle
Cattle-cattle transmission > shared mill equipment
Farm-farm > animal transport
Domesticated animals
Not highly pathogenic in humans, except immunocompromised
Still no sustainable human-human transmission
Surveillance is critical, especially in cattle
Hypothetical concern: If US farmer gets infected with both H5N1 and seasonal H1N1, this could lead to reassortment & create strain capable of human-human transmission
Avian influenza H7N9
Detected first in China
Infects both birds & humans
Limited person-person transmission occurred
RNA viruses & mutation rates
RNA viruses tend to have high mutation rates due to lack of proofreading of RdRp, leading to:
genetic drift & loss of immune recognition
reassortment (segmented genomes)
recombination, causing large-scale genetic change (genetic shift)
Explain overview of RT viruses
Viruses that use reverse transcription
Class VI (ssRNA-RT) = RNA is reverse transcribed into DNA
Class VII (dsRNA-RT) = Has gapped DNA genome that undergoes reverse transcription during replication
Reverse transcriptase
Enzyme that synthesizes DNA from RNA template
Highly conserved across species
Has catalytic site where nucleotide triphosphates are added to growing DNA strand
Not unique to viruses; exist in many organisms
Activities of RT
RdDp
Synthesizes DNA strand from RNA template
Primer-dependent
Produces RNA-DNA hybrid
RNase H / ribonuclease
Degrades RNA strand in hybrid
Endonuclease function
DdDp
Synthesizes complementary DNA strand, making a dsDNA
Broader importance of RT
RT is found in all domains of life
In humans, telomerase is a RT that solves the end-replication problem in linear chromosomes; carries an RNA template that extends 3’ ends of DNA to prevent shortening
Retrotransposons = fossil evidence of RT activity in genomes
Present in archaea, supporting idea of UCA
RT & RNA world hypothesis
RNA preceded DNA
RNA = both catalytic & information functions
RT may have bridged RNA-based life to DNA-based life
RNA can catalyze its own replication & protein synthesis, e.g., ribozyme activity of rRNA in ribosomes
DNA became permanent genetic storage due to stability of double-stranded structure
Explain ssRNA-RT (Class VI); HIV
Structure
Enveloped virus
2 +RNA copies ver virion
Infects CD4+ T cells & macrophages
Entry
Binds to host receptors
Fusion protein mediates entry
Capsid uncoats in cytoplasm
RT
1st strand synthesis = uses host tRNA primer bound at pbs for RT DNA from RNA template
RT degrades RNA; synthesizes complementary DNA strand
End up with linear dsDNA with long terminal repeats imported to nucleus for integration
Integration
Integrase inserts viral DNA into host genome
Permanent / cannot be excised
Can now be transcribed to produce
Viral proteins
RNA genome
Transcription & assembly
Alternative splicing produces different regulatory/accessory proteins unique to HIV
Common
Gag = capsid protein
Pol = RT+integrase fusion
Env = env protein
Gag-Pol fusion protein may form
Maturation
HIV are not infectious right after budding
Proteolysis of polyproteins are required postbudding to mature
Proteases inhibitors = target of HIV therapy
Why is HIV diploid?
2 RNA genomes linked via kissing loop complex
Only 1 RNA template is used for RT
Template switching allows
Recombination during DNA synthesis
Increased diversity
High mutation + high recombination = highly diverse virus population within 1 host
Molecular steps in RT
Host tRNA primer binds near 5’ end; extension space is short
Extension occurs, switching to 3’ end due to sequence similarity
Purine-rich tract (from degraded RNA) serves as primer for +strand synthesis
Template switching again as synthesis proceeds
End up with linear dsDNA with LTRs > promote integration
Consequences of integration
U now have permanent viral DNA in host genome
Retrovirus can be passed onto next generation if it infects germ cells > endogenization
Endogenization = provirus becomes part of germline DNA
Over time, they lose their ability to replicate
Endogenous retroviruses = ancient, non-replicating viral DNA
Retrotransposons = ancient RT elements, lacking envelop and cant form virions
42& of our genome = retroviral
Disease & oncogenesis
If integration occurs near cell cycle genes, this may activation oncogenes > tumor formation (Rous Sarcoma, Murine Leukemia)
HIV progression & therapy
Initially = high viral load + sharp CD4 drop
Latency period follows
AIDS onset occurs after gradual CD4 depletion
Antiretroviral therapy
Reduce viral production
Prevent CD4 depletion
Prevent immunosuppression
HIV drug targets ripf
Reverse transcriptase
Integrase
Protease
Fusion protein
Explain dsRNA-RT (Class VI); Hepatitis B virus
Structure
Enveloped virus with partial dsDNA genome (gapped)
+ strand = incomplete, linked with short RNA with 5’ cap
- strand = covalently linked to viral polymerase
Why genome is gapped
RT happens inside virion after capsid is closed during assembly
RT begins after packaging and does not finish > gapped dsDNA
Entry
Viral entry > import to nucleus
Host enzymes in nucleus repair/seal gap, forming relaxed circular DNA > template for transcription
Vesicle production in HBV infection
Infected cells release many non-infectious vesicles that display surface proteins of virus but do not contain genome
This outnumbers infectious vesicles, distracting immune system and assisting in immune evasion
Explain viroid as subviral element
Structure
Only naked RNA, circular, single-stranded, noncoding
Does not code any proteins
High secondary structure, which makes them stable & capable of ribozyme-like activities
2 families
Pospovirioidae
Rod-like shape
Replicates in nucleus
Has central conserved region
Avsunviroidae
Hammerhead ribozyme (self-cleaving structure)
Replicates in chloroplast
Transmission & replication
Entry: mechanical damage (plant wounds)
Spread: cell to cell via plasmodesmata; long-distance via phloem
Transmitted vertically thru seeds/pollens; horizontally
Replication mechanism
RNA → RNA via RdRp from host
DdRp
Example
Potato spindle tuber viroid – causes stunted, spindle-like potato plants
Coconut cadang cadang viroids: 246 nucleotides → encodes 0 genes
Explain satellites & satellite viruses as subviral element
Subviral agents that require “helper virus” for replication
Cannot replicate or form capsids independently
2 forms
Satellite nucleic acids (NA)
Do not encode capsid proteins
Instead, they’re packaged inside helper virus’ capsid
Some encode nonstructural proteins
Satellite viruses
Have their own capsids (complete virions)
Still require helper virus for replication enzymes
Difference from defective viruses
Defective viruses have homology with parent virus; they’re mutated forms of original viruses
Satellites are unrelated to helper virus genomically
e.g.,
Satellite = phage P4
Helper = phage P2
Satellite = AAV
Helper = Adenovirus
Explain AAV as subviral element
Naturally dependent on adenovirus for replication
Without adenovirus, it will be integrated into host genome
Upon future adenovirus infection, integrated AAV will be rescued & resume replication
Gene therapy
AAV is engineered
Replication genes replaced with transgenes
Helper functions are provided in packaging,
making AAV an efficient vector for gene delivery
Explain virophages as subviral element
Infect protist hosts and require co-infection with giant viruses (NCLDV – nucleocytoplasmic large DNA viruses)
Examples:
Helper Virus: Mamavirus, CroV
Virophage: Sputnik, Mavirus
Key Features
Replicate in host cytoplasm
Do not replicate inside helper virus, but depend on its transcription machinery
Reduce replication efficiency of the helper virus → beneficial to host survival
Evolutionary Implications:
Related to DNA transposons like Polintons/Mavericks
Possibly originated from virophages integrated into host genomes
These elements can be vertically transmitted, leading to diversity and expansion in host genomes
Explain Hepatitis Delta Virus as subviral element
Small circular RNA virus requiring Hepatitis B Virus (HBV) as helper
Co-infection with HBV leads to more severe disease
Like viroids, it is RNA-only and doesn’t encode a polymerase
Uses the envelope and coat proteins of Hepatitis B in order to complete its replication cycle
Current Mystery:
Other delta-like agents have been found via transcriptomics in different species
Their helper viruses are unknown or possibly not required
Explain prions as subviral element
Discovered by Stanley Prusiner
Protein-only infectious agents—no nucleic acid
Cause neurodegenerative diseases called Transmissible Spongiform Encephalopathies (TSEs)
Mechanism of Action
Normal prion protein (PrPᶜ) changes conformation into disease form (PrPˢᶜ)
PrPˢᶜ is stable, resists degradation, and induces others to misfold
Leads to accumulation of amyloid plaques in the brain → neurodegeneration
No immune response is mounted
Diseases
Scrapie – first TSE in sheep
Bovine Spongiform Encephalopathy (BSE) – Mad cow disease, spread via contaminated feed
Kuru – in humans due to cannibalism
Creutzfeldt-Jakob Disease (CJD) – human TSE; variant CJD (vCJD) linked to BSE
Types of TSEs
Infectious
Familial (inherited mutation in prnp)
Sporadic (random conversion to PrPˢᶜ)
Transmission & Entry
Prions can cross epithelial barriers
Taken up in gut (Peyer’s patches) and transported to the brain
Current Concern (USA):
Chronic Wasting Disease in cervids (deer family) linked to prion-contaminated soil