Genetics 3rd Midterms/Developmental Genetics
1/51
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
52 Terms
Development is controlled by what 2 types of protein coding genes?
Transcription Factor genes - function
Intercellular Signaling genes (ligand & receptor) - communication
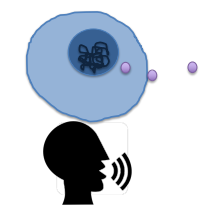
Intercellular Signaling Ligand genes
Encode secreted protein ligands that allow cells to talk to other cells (words/mouth of the cell)
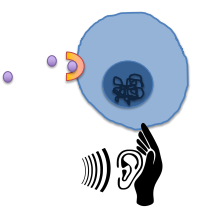
Intercellular Signaling Receptor genes
Encode cell surface receptor proteins that allow cells to listen to other cells (ears of the cell)
Transcription Factor genes
Encode TFs proteins that determine which other genes are turned on or off in a cell
Determine a cells function
How do cells “respond” to each other?
By changing expression of genes like TFs
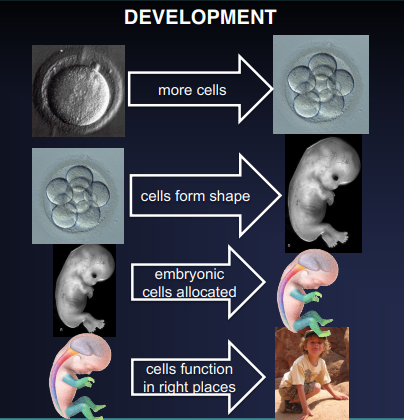
What are the 4 steps of Development?
Growth
Morphogenesis
Patterning
Differentiation
Cell Growth
1 cell becomes millions, billions, or trillions of cells through mitosis
Growth Rate
Early development: most cells divide at similar maximum rate
Later development/adulthood: different cell types divide at different rates
This allows organs/tissues to grow at different rates to achieve different sizes
Cell Cycle Checkpoint Proteins
The expression of these proteins in each cell determine the cell’s division rates
During development these genes are regulated by intercellular signaling receptors + ligands and TFs
What is necessary for anything to change size?
Impossible without additional nutrients and energy
Morphogenesis
The process by which the mass of embryonic cells “creates form” or “takes shape” of the organism or its parts (limbs, organs, etc.)
What drives morphogenesis?
This is mainly driven by morphogenetic movements which are the coordinated movements of embryonic cells relative to each other

Invagination
Morphogenetic movement in which the indenting of a sheet of cells forms a groove, tube, or cavity
Driven by changes in cell shape caused by changes in the cytoskeleton (cell skeleton)
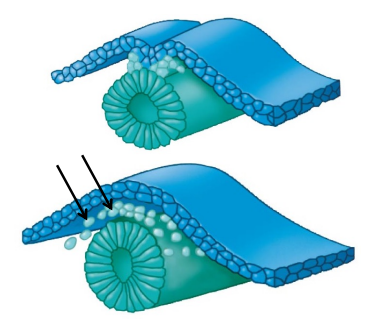
Migration
Amoeba-like movement of cells to a new place
Caused by changing expression of cell adhesion and cytoskeletal genes
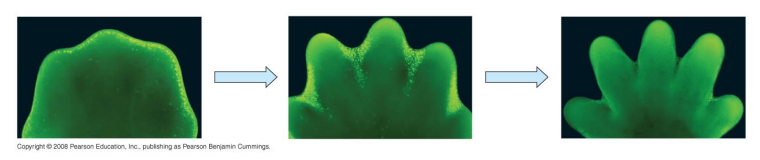
Apoptosis
Programmed cell death
Contributes to morphogenesis by removing cells and “sculpting” the body
Axial Patterning
The process by which cells in a developing embryo acquire TF combos that determines their position along an axis so they can develop into the correct structures in the right locations
Tissue Patterning
Where individual cells in the embryonic region progressively aquire their own specific TF combos
Maternal Determinants
Proteins and RNAs that are anchored to particular parts of the cytoplasm in the fertilized egg (zygote) that shape early embryonic development, inherited from mom. Bc of this, embryonic cells instantly have different combos of TFs without any new transcription or translation
Morphogen Gradients
Concentration gradients of secreted intercellular signaling ligands that pattern the embryo
Ligands
Often are maternal determinants or turned on by them. They diffuse across embryo btw cells and are received by cells with the corresponding receptor. Different concentrations of these cause responding cells to express different kinds and amounts of TFs, “patterning” them
Asymmetrical Cell Division
A mini version of maternal determinants
A type of cell division where a parent cell divides to produce two daughter cells with different fates—meaning they differ in size, components, gene expression, or developmental potential
Short-Range Intercellular Signaling
A mini version of morphogen gradients, cell-to-cell discussion. Tells cells to morph by changing active TFs, i.e. eye cell to bone cell
Differentiation
Process that embryonic cells mature and take on their final form and function. During this, TFs turn on hundreds of tissue-specific effector genes, allowing a mature cell to do its job
Main Stages of Vertebrate Development
Zygote
Cleavage Stage
Blastula
Gastrula
Neurulation
Organogenesis
Zygote
Fertilized egg (diploid)
Cleavage Stage
Ball of cells created by repeated division of the zygote, growth, patterning
Blastula
Ball of cells becomes hallow, cells move to edge of ball to create open space
Gastrulation (gastrula)
The blastula invaginated to create 2 cell layers. The outer is called ectoderm, which differentiates into the spine and nervous system. The inner layer later divides to become the mesoderm and endoderm.
Neurulation
Formation of the nervous system by rolling up the dorsal (top) of the ectoderm into a neural tube. Failure results in spina bifida - the spinal cord is still open outside the spine and the skin
Organogenesis
Development of organs and other structures by coordinated growth, patterning, differentiation, and morphogenesis
Substitution Mutation
Where one nucleotide replaces another in the sequence. Rarely results in anything except the extension (removal of a stop codon) or shortening of a genetic sequence (addition of a stop codon).
Causes of Substitution Mutations
DNA polymerase errors not fixed by proofreading - 1 out of 10 billion bps is missing
Spontaneous deamination of “C” cytosine turns to “U” uracil
Alkylating chemicals like ENU and EMS change the structure of N-containing bases
UV light can create substitutions indirectly via DNA repair mechanisms
Consequences of Substitution Mutations
In protein-coding exons, single amino acids can be replaced by another; the sequenced can be shortened, extended, or have no effect
In non-protein-coding sequences left in to be read as protein-coding sequences, there will statistically be a stop codon read inside it before the real protein-coding STOP codon, shortening it
Loss of splice donor/acceptor sites
Interferes with splicing, often leading to introns not being removed and signaling a STOP codon before the protein-coding sequence
Substitution Mutations Effect on Cis-Regulatory Elements
There is little to no effect on gene expression because TFs don’t need exact binding site sequences, each type of TF binds to several sites in an enhancer, and enhancers/silencers don’t follow the genetic code
Indels
Insertions/deletions of one or more nucleotides into a DNA sequence. Most are 1-10 nucleotides, but can be 1000s of bps or entire arms of chromosomes
Replications Slippage
The cause of indels
Consequences of Indels
Generally, few for small non-protein-coding sequences. In protein-coding exons, it can disrupt open reading frames, causing frameshift mutations possibly creating early STOP codons
Frameshift Mutations
Insertion or deletion of nucleotides in a DNA sequence, shifting the reading of the code by 1, 2, or 3 frames positively or negatively. Often results in a completely different, usually non-functional, protein. Single or double bp indels shift all reading frames whereas triple bp indels do nothing unless it is a STOP codon
Open Reading Frames (ORFs)
Long stretches (hundreds of bps) without STOP codons except for the very last codon. In all exons and in no CREs. Do not have to start with a START codon, but always end with a STOP codon
Plyotropy
When a single gene influences multiple, seemingly unrelated traits in an organism
Chromosomal Mutations
Insertions and deletions on a massive scale, often affecting dozens of genes. Can occur btw parts of the same chromosome, btw maternal and paternal homologous chromosomes, and btw non-homologous chromosomes. Requires breaking and rejoining of chromosomal DNA
Chromosomal Mutations consist of
Deletions, duplications, inversions, and translocations
Causes of Chromosomal Mutations
Intrachromosomal “crossing over” (recombination) during meiosis possibly leading to duplications and deletions
…
Consequences of Chromosomal Mutations
Changes in gene expression from new/missing CREs
Creation of truncated proteins by causing frameshifts or exon deletion
Creation of new proteins with new functions by duplicating exons or combining exons from different genes
Crossing Over
Chunks/sections of chromosomes are swapped btw strands during meiosis
Truncated Proteins
Incomplete or shortened versions of proteins resulting from genetic mutation. Can introduce premature STOP codons, leading to the production of a protein lacking in essential function, rendering it nonfunctional or unstable
Chromosome Number Mutations
Changes in the number of chromosomes in the gene from n. N = number of chromosomes normally found
Aneuploidy
Number of chromosomes is not exactly a multiple of n, ±1 or ±2 chromosomes
Aberrant Euploidy
Number of chromosomes is an exact multiple of n that is not typical, i.e. double or triple the amount of chromosomes