UNIT 2 microbiology test final set
1/89
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
90 Terms
Gene Expression
The process of converting DNA information (a gene) into a functional protein that dictates the cell's phenotype.
Operon
A cluster of genes controlled by a single promoter. Produces a polycistronic mRNA that encodes multiple proteins.
Regulon
Multiple operons (or single genes) that are spread throughout the genome but are all regulated by a single regulatory protein.
Transcription Control
Regulation at the level of DNA→mRNA synthesis. This is the most efficient way to save energy/resources.
Repressor Protein
A DNA-binding protein that blocks RNA polymerase from binding to the operator. Used in negative control (e.g., Arginine and Lactose operons).
Inducer vs. Co-repressor
Inducer: A substrate (e.g., lactose) that turns ON a gene by binding the repressor and freeing the operator.
Co-repressor: An end product (e.g., arginine) that turns OFF a gene by binding the repressor and allowing it to bind the operator.
Activator Protein
A DNA-binding protein that is required for RNA polymerase to bind. Used in positive control (e.g., Maltose operon).
Catabolite Repression
The mechanism that ensures glucose (the preferred carbon source) is used first. It relies on low glucose → high cAMP → CRP activator binding to turn ON other catabolic operons.
Coupled Transcription/Translation
Occurs only in prokaryotes. Ribosomes begin translation of the mRNA while RNA polymerase is still performing transcription.
Post-transcriptional Reg.
Control mechanisms acting on the mRNA or protein level. It provides a quicker response than regulating transcription. Examples: sRNA, Riboswitches, Allosteric control.
Riboswitch / sRNA
Riboswitch: mRNA binds a small metabolite, forcing it to fold into a loop that blocks the ribosome
. sRNA (small RNA): A non-coding RNA molecule that base-pairs with mRNA to block translation or trigger destruction.
Allosteric Regulation
The fastest regulatory mechanism. An end product (effector) binds to an allosteric site on an enzyme, changing the active site's shape to block the substrate from binding and immediately stopping the pathway.
Point Mutations
Changes involving a single base pair. Examples: Silent, Missense, and Nonsense mutations.
Frameshift Mutation
Insertion or deletion of 1 or 2 bases within a coding region. This shifts the reading frame for all subsequent codons.
Reversion Mutation
A second mutation that restores the wild-type genotype or phenotype. Can restore a misread codon or re-establish a correct reading frame.
Homologous Recombination
The exchange of a DNA segment from one molecule to another where the segments have nearly the same sequence. This process is required to make most HGT events heritable.
Transformation
Horizontal transfer via the uptake of naked, free donor DNA from the environment by a recipient cell. The cell must be competent to take up the DNA.
Transduction
Virus-mediated transfer of donor DNA to a recipient cell. Examples: Generalized (random gene) and Specialized (adjacent gene).
Generalized vs. Specialized Transduction
Generalized: Random pieces of chromosomal DNA are accidentally packaged into a lytic phage head.
Specialized: Incorrect excision of a lysogenic prophage results in the phage carrying a small, adjacent piece of host DNA.
Conjugation
Direct-contact transfer of DNA from a donor to a recipient cell, typically mediated by a conjugative plasmid.
F+ vs. Hfr
F+ cell: Donor cell with the F plasmid existing separately from the chromosome.
Hfr cell: Donor cell with the F plasmid integrated into the chromosome.
Plasmid
An extrachromosomal DNA element that replicates independently of the host chromosome. Often encodes non-essential genes like antibiotic resistance.
F Plasmid Components
oriT (Origin of Transfer): Where DNA strand is nicked to begin transfer.
tra region/Pilus gene: Encodes the protein tube (pilus) that facilitates contact and transfer.
IS sequences: Allow the plasmid to integrate into the chromosome to form an Hfr cell.
Phage Life Cycles
Lytic Cycle: Virus replicates immediately, resulting in lysis of host cell.
Lysogenic Cycle: Virus DNA integrates into host DNA (prophage) and replicates with the cell without lysis.
Phylogeny
Evolutionary relationships of organisms, visualized as tree diagrams.
Molecular Clock
The concept that DNA sequences accumulate mutations over time. Used to measure evolutionary distance. 16S rRNA is widely used as a reliable molecular clock.
Phylogenetic Tree Components
Root: Most recent common ancestor of all organisms in the tree.
Nodes: Common ancestors where lineages diverge.
Branch Length: Proportional to evolutionary distance.
Clade: A group of closely related organisms.
16S rRNA
The gene encoding the small subunit ribosomal RNA. It is used for phylogeny because it performs the same function in all organisms and accumulates mutations slowly enough to track evolution.
DNA Alignment
The first step in building a tree. Sequences are arranged to find overlaps and mismatches (differences).
Distance Matrix
A table calculated from aligned sequences that shows the number of sequence differences between every pair of organisms. This matrix is used by software to build the tree.
Microorganism Habitat
The environment that provides favorable conditions for growth, including nutrients. Examples: Soil, lakes, GI tract.
Microenvironment
Smaller habitats where microorganisms are found (e.g., a pore space in a soil aggregate).
Niche
The resources utilized by a microorganism. It is defined by a combination of biotic and abiotic factors.
Population vs. Guild
Population: Individual cells reproducing
. Guild: Populations that do similar or related metabolism (e.g., denitrifiers).
Community
Populations/Guilds that live together (e.g., in a lake stratum).
Culture-Independent Methods
Methods used to analyze a microbial community directly from a sample without growing it in the laboratory.
Soil Aggregate
Groups of soil particles (sand, silt, clay) bound together. They contain diverse solid, liquid, and gas phases that create many microenvironments.
Anoxic Microenvironment
In a soil aggregate (or biofilm), the interior may become anoxic because O2 diffusion is inhibited and O2 is consumed by outer-layer microbes. Anaerobic metabolism (fermentation) then takes over.
Rhizodeposition
The process where plants release (excrete) organic carbon compounds (sugars) into the soil from their roots. This carbon acts as a food source for soil microbes.
Nodule Formation (Signals)
Flavonoids (from root) signal rhizobia. Rhizobia respond by synthesizing Nod Factors. Nod Factors induce root hair curling , allowing the bacteria to enter via the infection thread.
Bacteroid
Pleomorphic bacterial cells inside the root nodule that are specialized for fixing nitrogen.
Nitrogenase / Energy
The enzyme system that performs nitrogen fixation (N2→NH3). This reaction requires a large amount of ATP. The ATP is supplied by the aerobic respiration of the bacteroids.
Leghemoglobin (Lb)
A protein produced by the plant that binds oxygen (O2). It transports O2 for aerobic respiration but also protects the oxygen-sensitive nitrogenase enzyme.
Human Microbiota
All the microbes living in and on the human body. The main sites are the intestines (largest population), mouth, and skin.
Sterile Sites
Areas that normally have no microbes. Examples include the blood, lower respiratory tract (lungs), and bladder.
GI Tract Gradient
Microbial diversity is low in the Stomach (pH 2) and increases massively in the Colon (pH 7). The Colon is overwhelmingly anaerobic.
Symbiotic GI Functions
Microbes provide the host with essential: Vitamin synthesis (B12,K), Glycosidase activity (digestion of complex food), Steroid metabolism, and Antagonism of pathogens.
Lactobacillus acidophilus
A beneficial microbe in the vagina. It ferments glycogen to lactic acid, creating a low pH (∼5) environment that inhibits pathogens (bacterial interference).
H. pylori Survival
H. pylori uses the Urease enzyme to convert urea into ammonia (NH3). Ammonia neutralizes the stomach acid in the local microenvironment, allowing the bacteria to colonize.
Dental Biofilm
Initial formation is attachment to the tooth's protein pellicle (using adhesins). Microbes ferment sucrose to make dextran (a glucose polymer) via dextransucrase, which aids attachment to form thick plaque (biofilm).
Ruminant Digestion
Ruminants lack enzymes to digest plant fiber (cellulose). Microbes in the Rumen ferment the fiber into Volatile Fatty Acids (VFAs), which the host absorbs for energy.
Three main categories of methods for controlling microbial growth?
Growth inhibition by environmental conditions
Killing/reduction in numbers using physical methods (high heat, radiation, filtration)
killing/reduction using non chemotherapeutic chemicals
Decimal reduction time
The time (or dose of radiation) required to achieve a 10 fold reduction in viable number of organisms
purpose and operating condition of an autoclave
purpose- kill all microbes and spores
Operating condition- 121 C at 15lb/in²
what is the purpose of pasteurization
reduce the number of viable microbes but not kill all of them
used to increase storage life
autoclave would be to harsh and destroy food

Batch method
63C for 30 min
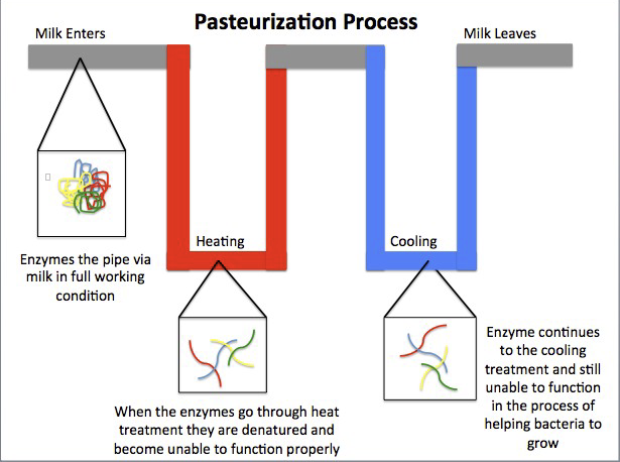
continuous flow method
72 C for 15 seconds follows by cooling to 4 C
Sterilizing effect and primary use of Uv light for microbial control
Sterilizing effect- Primarily causes damage to DNA and RNA
Uses- disinfecting surfaces, air, and water
Sterilizing effect and primary use of Gamma Rays (ionizing radiation)
sterilizing effect: causes damage to DNA, RNA, and proteins
uses: Sterilizing medical devices, foods, and pharmaceuticals
Filtration in liquids and gases
membrane filters use uniform sized holes to remove microbes from liquids 0.2 microns
HEPA filters remove 99.97 particles using 0.3 micron holes
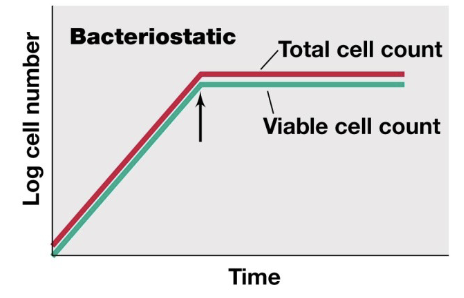
bacteriostatic
stops bacterial growth
total cell count plateaus, viable cell count plateaus
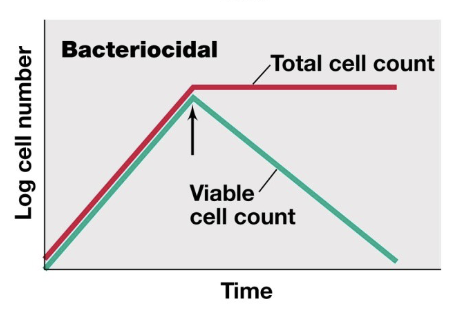
Bacteriocidal
kills the organism
total cell count plateaus, viable cell count decreases
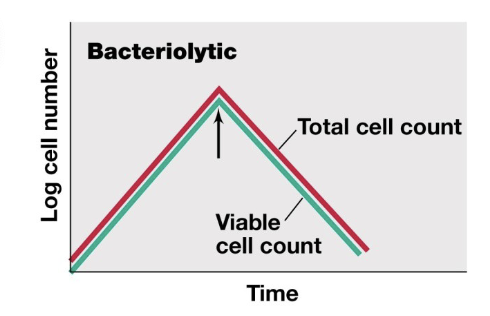
Bacteriolytic
kills by lysis
total cell count decreases, viable cell count decreases
Sterilizers
kill all microbes and spores
toxic to animals
used: hospital labs, instruments, and medical devices
Disinfectants
used on non-living surfaces
may kill spores
less toxic
use: homes, hospitals
Antiseptics
nontoxic to living tissues
use: wounds directly on the body
Sanitizers
REDUCE number of microbes
low toxicity to animals
use: food prep
Antimicrobial agent factors
concentration
exposure time
addition of other chemicals
Function of the genome
to store all the genetic information of a cell, including genes an operons, encoded in the sequence of DNA
information storage computer vs cell
computer: 1 and 0 s (multiples of 6)
cell: GATC (multiples of 3)
Information retrieval: computer vs cell
computer: Retrieves info into RAM with files
Cell: retrieves info by transcribing genes into mRNA
Executing instructions: computer vs cell
Computer: runs programs
cell: translation of mRNA to run metabolic pathways
Information obtained from genome sequencing
infer metabolic pathways
measure protein and RNA levels
detect genes involved in virulence
Use of targeted Sequencing
sequence a specific gene or region of a genome
often to compare that gene across many organisms or look at specific mutations
Use of whole genome sequencing
study all genes in a microorganism
necessary to discover regulation, pathways and metabolism
Open Reading Frame
A sequence of DNA that has a ribosome bonding site, start codon and an end codon
encodes for a protein
steps to analyze DNA sequencing data
align shared sequence readings into contigs
arrange contigs into scaffolds
Gap closure and finishing
predict genes (ORFs) and location
annotation- assign function to genes based on similarity to known genes
contigs
larger regions of aligned sequence data
bioinformatics
a field of biology that uses computational tools to store, analyze, and compare genome data
reason for smaller genomes in parasitic bacteria
can delete genes and metabolic pathways that host provides
what genome sequences reveal about uncultured organisms
they can reveal metabolic pathways, energy production mechanisms, nutrient requirements, and evolutionary relationships without needing to grow the organism in a lab.
Reason for using genome sequencing for most microbes
over 95% of microorganisms cannot cultured in a lab
Pathogenicity islands PAI
distinct clusters of genes on a chromosomes that are associated with an organisms ability to causes disease
Core Genome
The set of genes that are shared by all strains within a species or a group of compared organisms
Pan genome
The entire set of genes from all strains within a species or group
horizontal gene transfer
incorporating new genes from other organisms
vertical gene transfer
the transfer of genetic material from parent to offspring
Transcriptomics
The complete set of RNA transcripts that are produced by an organism under a specific set of conditions
proteomics
the large scale study of proteins their structures and functions
metabolomics
the scientific study of a set of metabolites present within an organism cell or tissue