Bio 219 Exam 2 WVU
1/84
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
85 Terms
Autocrine signaling
When a cell sends a signal to itself
Paracrine signaling
When a cell sends a signal to another cell that is close by
Juxtacrine signaling
Cell sends a signal to another cell directly through a channel/gap junction
Endocrine signaling
Cell sends a signal to another cell that is far away through the blood stream
Anti-apoptotic
A type of cellular response to a signal in which the cell continues it’s normal functions in order to survive
Growth factor
A type of cellular response to a signal in which the cell grows and divides into new cells
Differentiate
A type of cellular response to a signal in which the cell changes identites
Ligand
A molecule that acts as a cellular signal.
They include hormones and local mediators.
They bind to transmembrane protein receptors non-covalently, leading to an allosteric change and for the signal to be amplified
Highly specific to the receptors they bind to
What are things that act as cellular signals that are NOT ligands?
Heat
Light
Motion/force
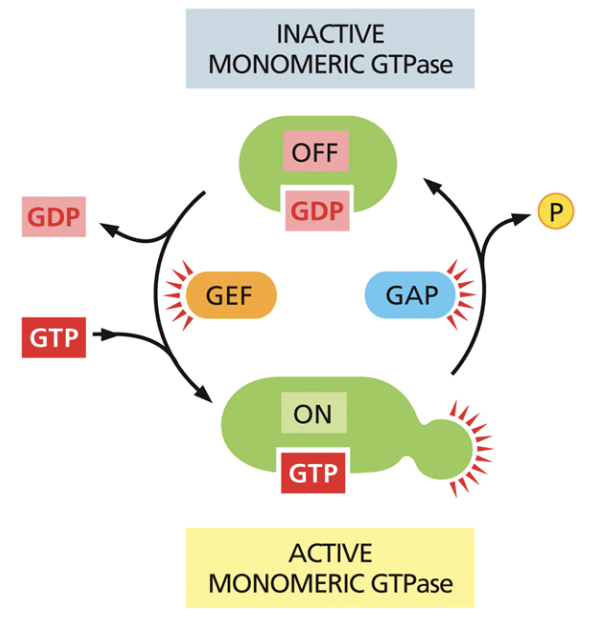
What does GTP do in signal transduction?
Regulates g-proteins by activating them through allostery
G-proteins are “on” when they’re bound to GTP and “off” when they’re bound to GDP
Guanosine exchange factor (GEF)
Makes GDP into GTP, which then binds to the protein.
Enables g-proteins to go from off —> on
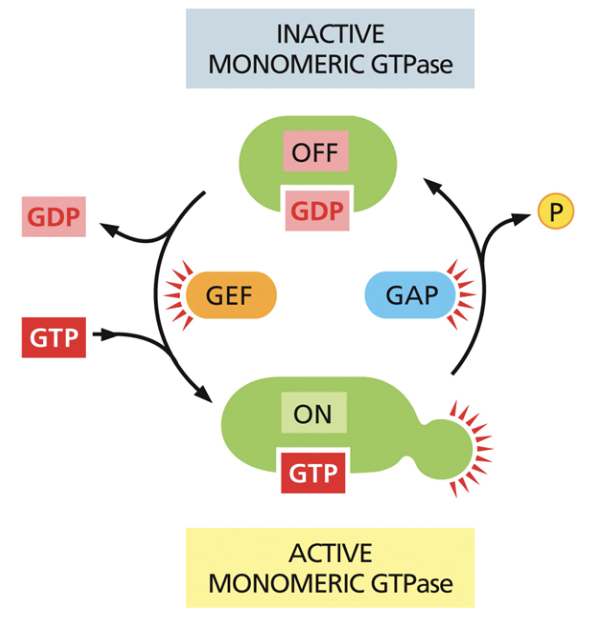
GTPase activating proteins (GAP)
Enables GDP hydrolysis, causing a phosphate to fall off and for GDP to bind to the protein.
Makes g-proteins go from on —> off
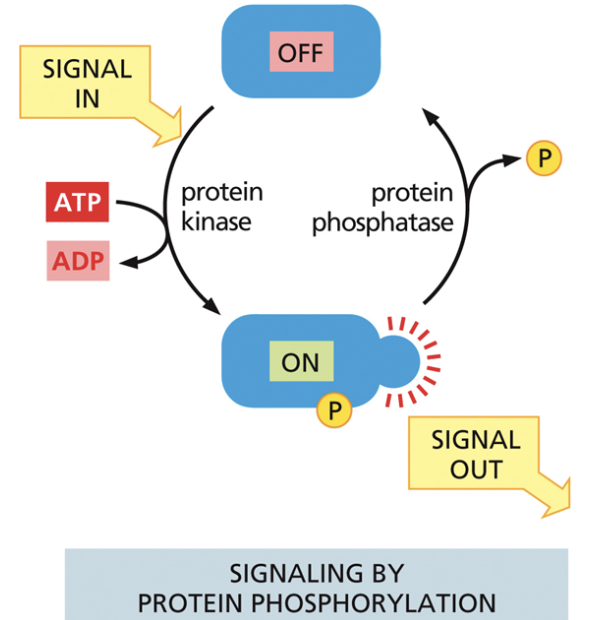
Kinase
Used in cell signaling. Moves phosphate from ATP to a recipient molecule
A molecule goes from off —> on when ATP becomes ADP and phosphate binds
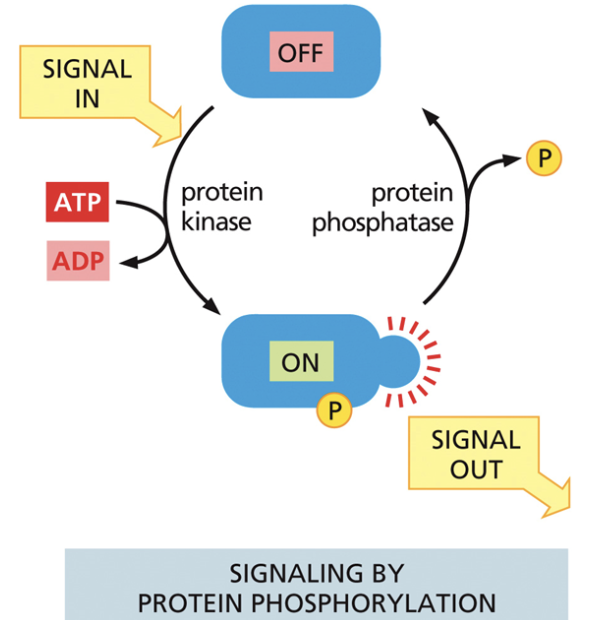
Protein phosphatase
Used in cell signaling. Removes the phosphate group from the molecule
Causes the molecule to go from on —> off
Ion coupled receptors
A common signaling mechanism
Opens channels when a ligand binds to a receptor and induces allostery
Channel closes when the ligand falls off
Provides ions for cell functions and creates action potential
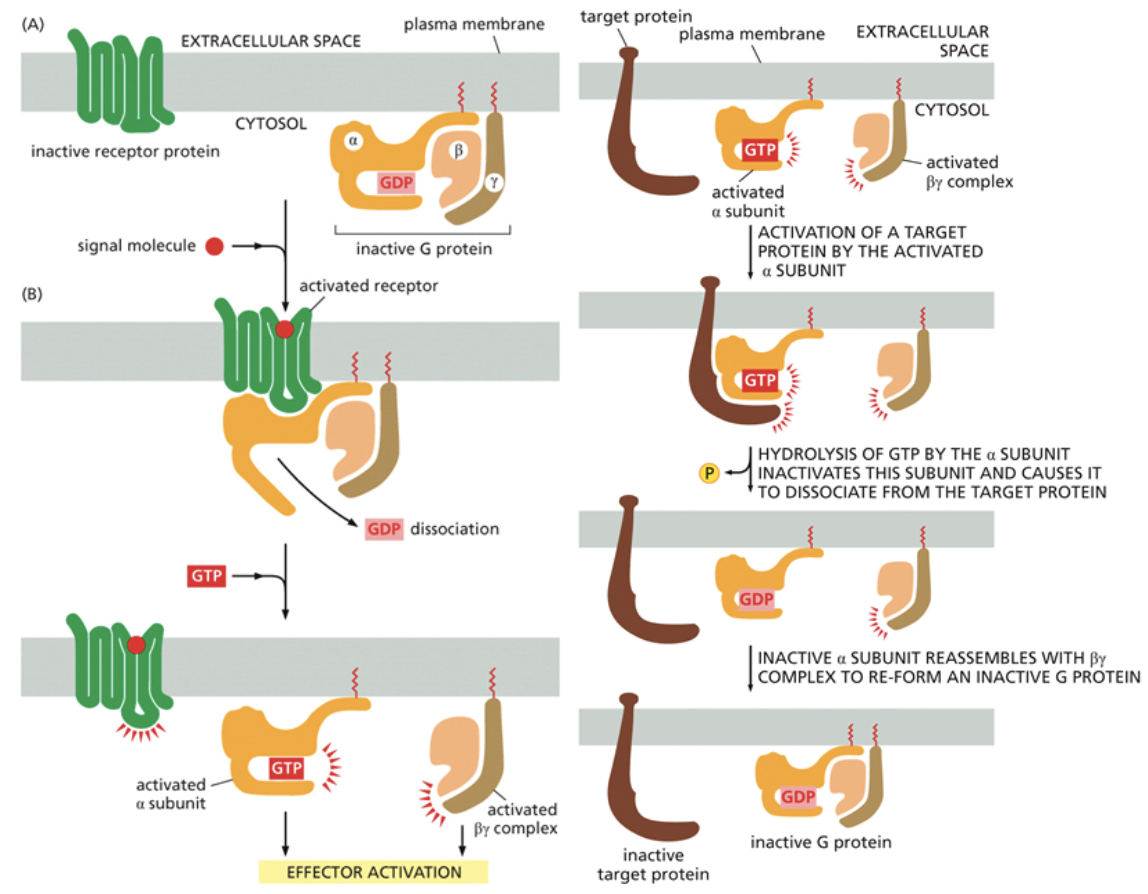
G-protein coupled receptors
A common signaling mechanism
Works with a g-protein inside of the cell to transmit a signal throughout. This g-protein is heterotrimeric, it has three distinct subunits
They are activated by the binding of GTP
Second messengers are the first molecules to leave the cell. Transmembrane proteins cannot leave the cell, but these messengers can
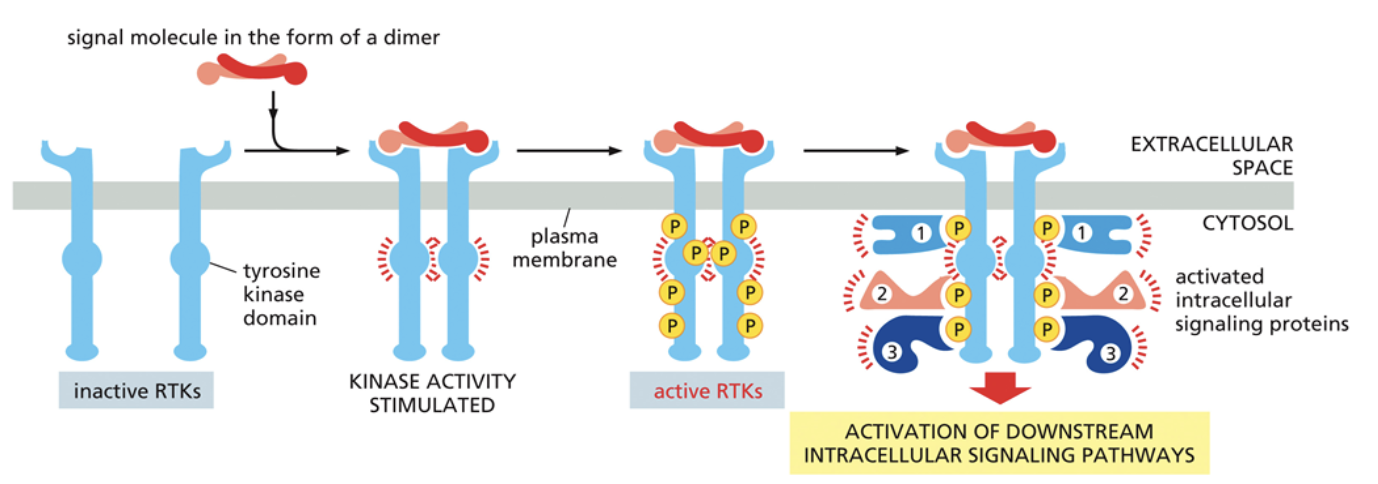
Receptor Tyrosine Kinase (RTK) receptors
A common signaling mechanism
Phosphorylate each other to transmit signals
Have three different parts: The ligand binding domain, which is outside of the cell and where the ligand binds; The transmembrane domain, which goes across the membrane; The enzymatic domain, which is a kinase that moves phosphates from ATP —> new molecules
RKT’s always act as a dimer that phosphorylates other parts of the dimer. Called transautophosphorylation. A ligand binding to the domain activates this. The right monomer phosphorylates the left monomer, and vice versa.
The phosphorylation of RKT essentially opens up new binding sites for different proteins. Often, the new proteins that bind are also kinases, which sets off a cascade of kinases activating
Regulatory region
A section of DNA that codes for where and how much of a product is made
Codon
Three nucleotides in a triplet that code for one amino acid
More than one codon can code for the same amino acid, it is degenerate
An individual nucleotide can only be apart of one codon, it is non-overlapping
Missense mutation
A nucleotide is replaced by another nucleotide
Nonsense mutation
A nucleotide is replaced by a stop codon
Frameshift mutation
A nucleotide is deleted, rather than replaced. This leads to every codon beyond the mutation being shifted by one.
Gene
A DNA sequence that contains information on how to make specific products. A coding region of DNA
Allele
A given gene with different variations due to being at different parts of the chromosome.
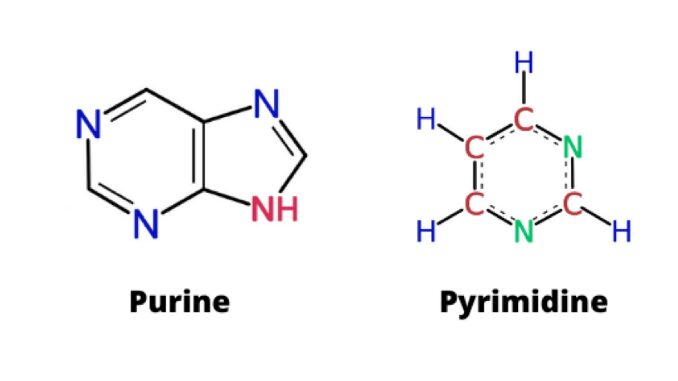
Purines
Types of nitrogenous bases in DNA and RNA that have a double ring structure.
Includes the bases Adenine and Guanine
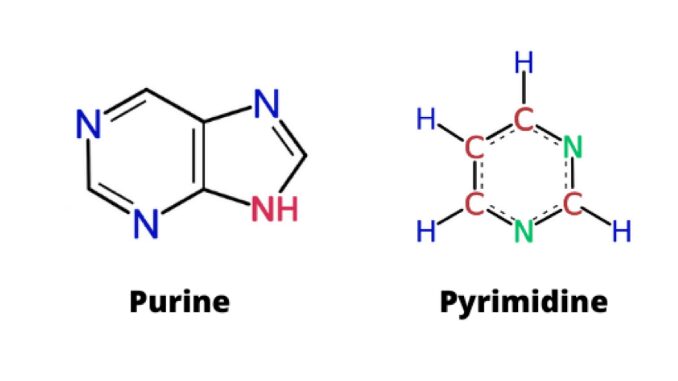
Pyrimidines
Types of nitrogenous bases in DNA and RNA that have a single ring structure.
DNA pyrimidines include the bases Cytosine and Thymine. RNA pyrimidines include Cytosine and Uracil
DNA polymerase
An enzyme that facilitates the condensation reaction that binds two nucleotides together
In this reaction, the 3’ hydroxyl group of one monomer is linked to the 5’ phosphate of another monomer. This creates covalent bonds along the backbone of DNA
Annealing
To recombine DNA into the double stranded form
Lamin
Connects the nuclear membrane to DNA
Changes to lamin structure coordinate nuclear membrane breakdown and reformation during mitosis
Major groove
The part of the DNA helix that goes in the front
Minor groove
The part of the DNA helix that goes in the back
Euchromatin
Bound by few proteins, so it’s less compact
Spaghetti like strands of DNA
More accessible to proteins that perform replication, transcription and translation
Genes that encode for important functions are typically in the Euchromatin region of the nucleus
Heterochromatin
Spaghetti like strands of DNA
Bound to more proteins than Euchromatin, so it’s more compact
Less accessible to proteins that perform replication, transcription and translation
Most DNA is in the form of heterochromatin
Nucleosome
A DNA and protein complex. DNA wraps tighter around histones
DNA portion of nucleosomes contains 75 base pair wrap arounds, wrapping around twice for a total of 150 base pairs
DNA is super long. Nucleosomes prevent DNA from breaking, taking up too much space or getting tangled
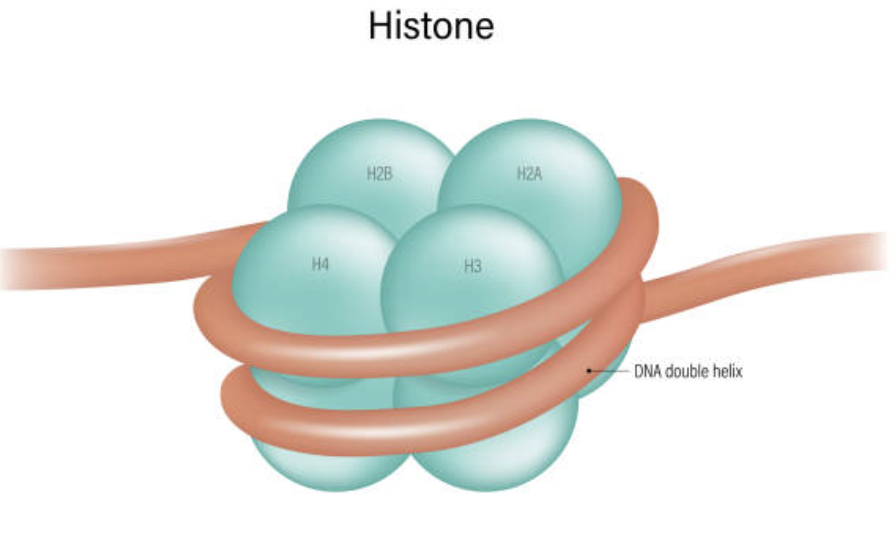
Histone
The protein DNA wraps around
There are four types of histones: H2A, H2B, H3, and H4
They exist in octets with 8 proteins
Describe recruitment in transcription
The first step of transcription
RNA polymerase is recruited to the correct part of the chromosome
DNA is targeted to the correct location by two kinds of factors: Cis factors, which are factors built into the same piece of DNA that RNA is trying to copy and trans factors, which are factors that bind to cis factors and were built in another location
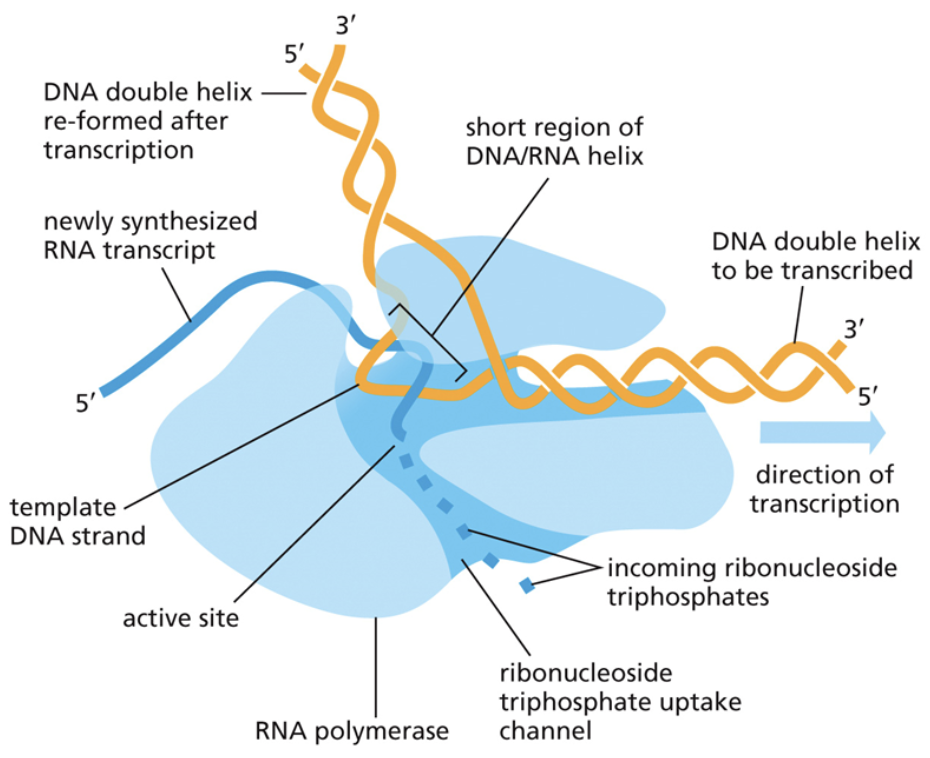
Describe initiation in transcription
Initiation is the second step of transcription
The DNA double helix opens, and RNA polymerase begins to read it
Describe elongation in transcription
Elongation is the third step of transcription
It is a continuation of initiation. The new RNA polymer gets longer and longer, adding each new part through condensation reactions
Uses the same base pair bonding as DNA, but with U instead of T
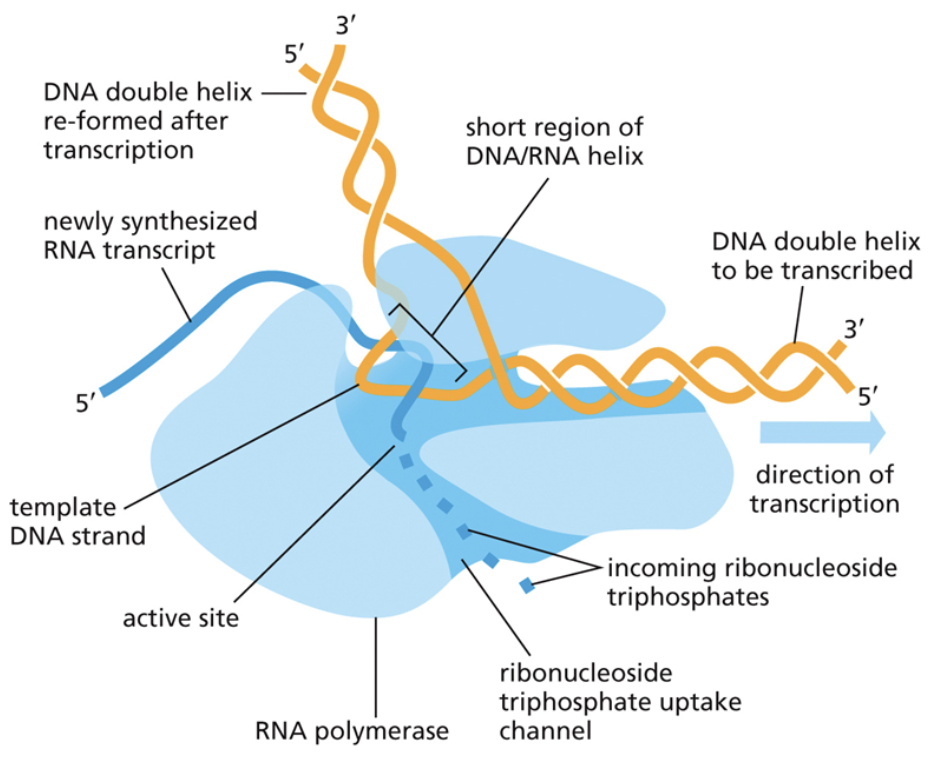
Describe termination in transcription
Termination is the final step in transcription
RNA polymerase reaches a STOP codon and ceases transcription
Cells use different types of signals to cease transcription
Rho dependent termination
A type of signal used to stop translation used by prokaryotes only
A protein called Rho recognizes a sequence in newly made RNA and runs up the strand until it reaches RNA polymerase
Once it reaches RNA polymerase, it unwinds the DNA-RNA hybrid inside RNA polymerase so transcription stops
Rho independent termination
A type of signal used to stop translation used by prokaryotes only
A bulky hairpin loop is made. It is made of an RNA strand binding to itself with G and C
When it runs into a sequence with lots of A and U bonds, the binding affinity between DNA and RNA weakens, and the DNA-RNA hybrid breaks apart, ceasing transcription
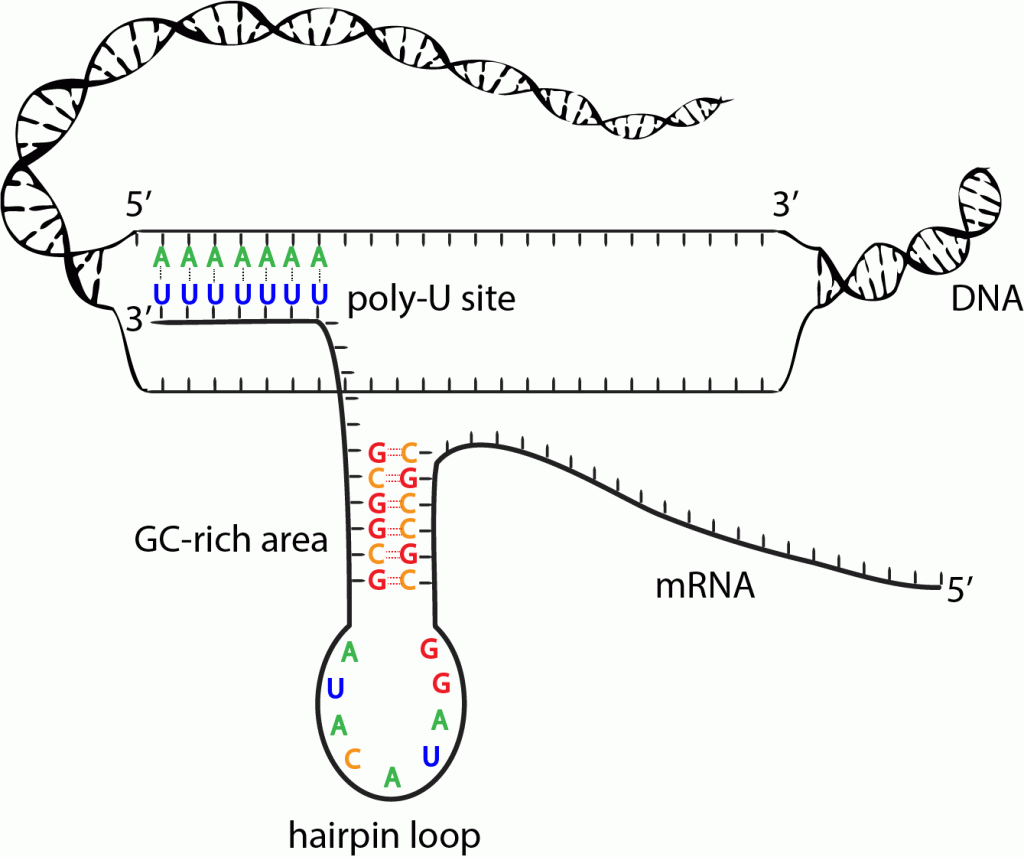
Cleavage
A type of signal used to stop translation used by eukaryotes only
RNA polymerase transcribes a STOP signal bound by a nuclease
Nucleases cut the RNA downstream of the STOP signal, which ceases transcription
Messenger RNA (mRNA)
Assembled by RNA polymerase II
Produced by transcription as a template for protein synthesis
mRNA must be processed. The processing of pre-mRNA includes the capping of it at the 5’ location, RNA splicing, and the addition of a polyA tail. This processing occurs in the nucleus
How and why does capping of mRNA occur?
Capping is the addition of a 7 methyl guanosine cap (7mG cap) at the 5’ end.
It is attached to the RNA transcript through a phosphate bridge in a 5’ to 5’ mechanism.
This bridge acts as a disguise for RNAse enzymes in the cytoplasm that want to consume it in case it is RNA from a foreign organism. This is because the phosphodiester bonds are at the 5’ to 5’ location.
RNAses recognize 5’ to 3’ phosphodiester bonds, leaving capped RNAs in the clear
How and why are tails added to mRNA?
mRNA is protected at 3’ end by a polyA tail. Tails are made by a series of ATPs
RNAses also want to degrade mRNA at the 3’ end. When A’s are added, it creates a competition between the addition of A’s and the removal of A by RNAses.
The longer the polyA tail, the longer it will take RNAses to degrade the coding, important parts of mRNA.
The polyA tail is initially added in the nucleus, but additional A’s can be added in the cytoplasm after nuclear export
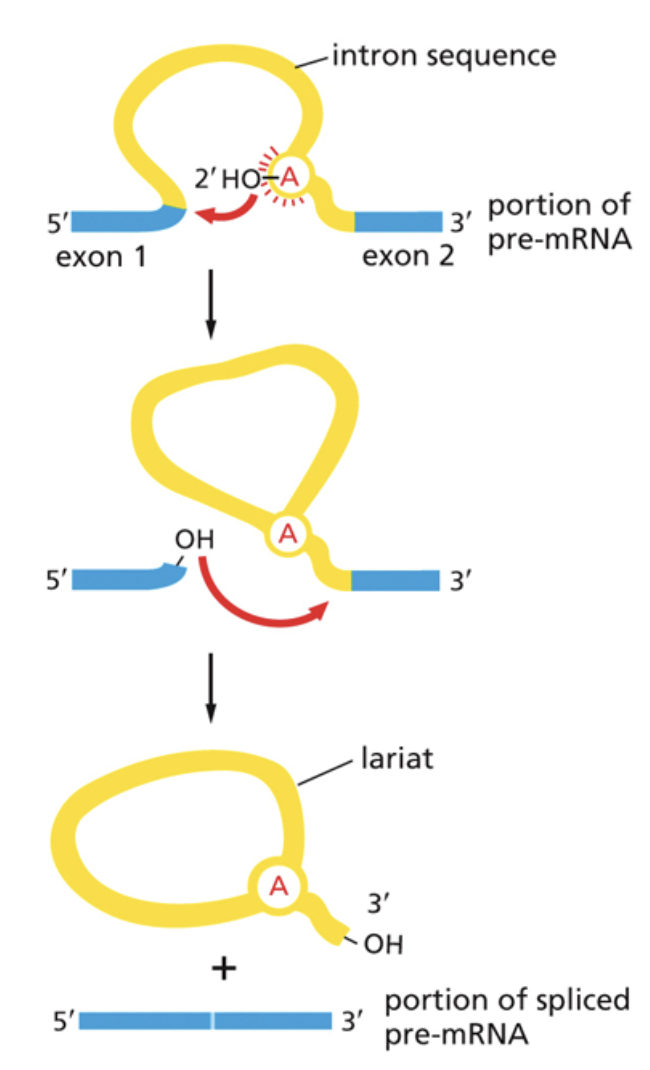
How and why does splicing occur in mRNAs?
Some non-coding sections of mRNA, introns, are removed via splicing. Exons are left in the code and expressed
Splicing occurs in two steps:
1. The free 3’ OH group attacks the 5’ splice site of exon 1 and cuts it. This releases the 5’ OH group, leaving it free to attack the 3’ splice site. It also creates the lariat
2. It then creates a new phosphodiester bond between the 3’ OH group and the 5’ phosphate of the second exon.
Splicing is regulated by SnRNPs. They use RNA as a catalyst.
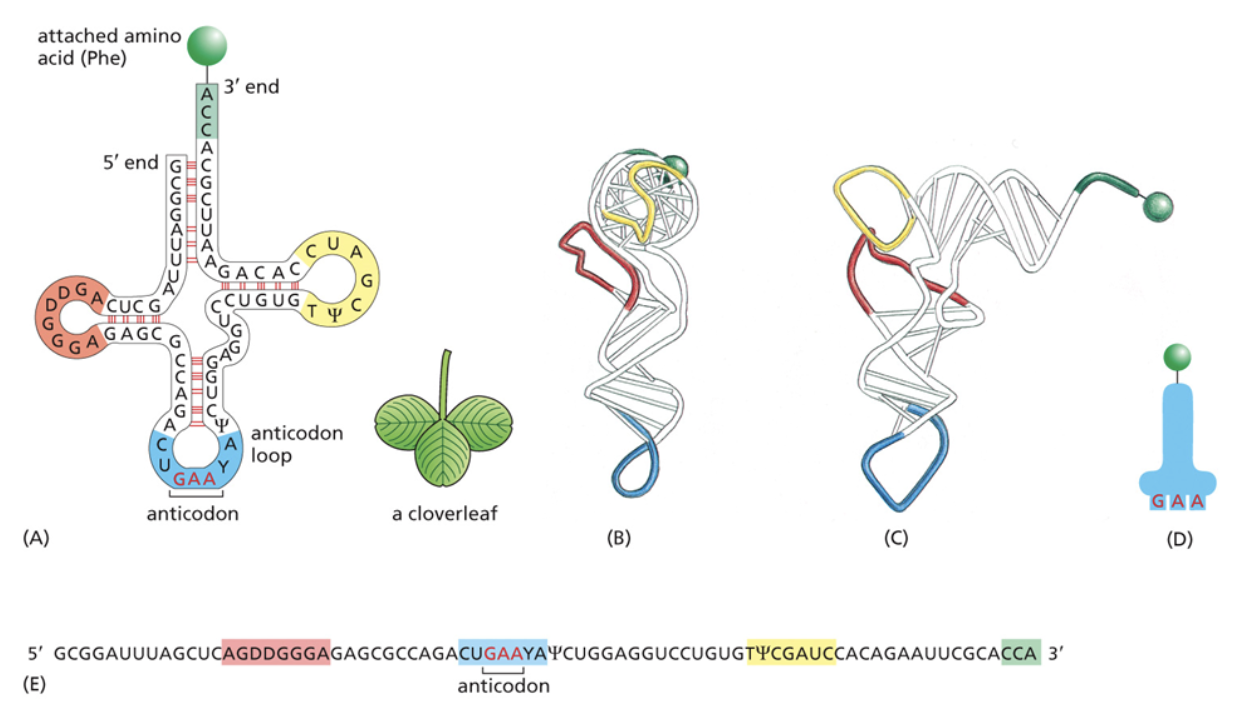
Transfer RNA (tRNA)
Assembled by RNA polymerase III
Act as a translator between mRNA and amino acids during protein synthesis
pre-tRNA goes through a lot of processing to become functional tRNA. This processing occurs after transcription is terminated but before tRNA is exported out of the nucleus.
After processing tRNA is later connected to an amino acid and folds into a secondary structure, then folds again to form the tertiary structure that makes an L shape.
At the bottom of the L shape, there is the anticodon that will be bound to the mRNA during translation
On the opposite side of the anticodon, there is the an amino acid that allows it to connect to other amino acids in the chain when RNA is translated
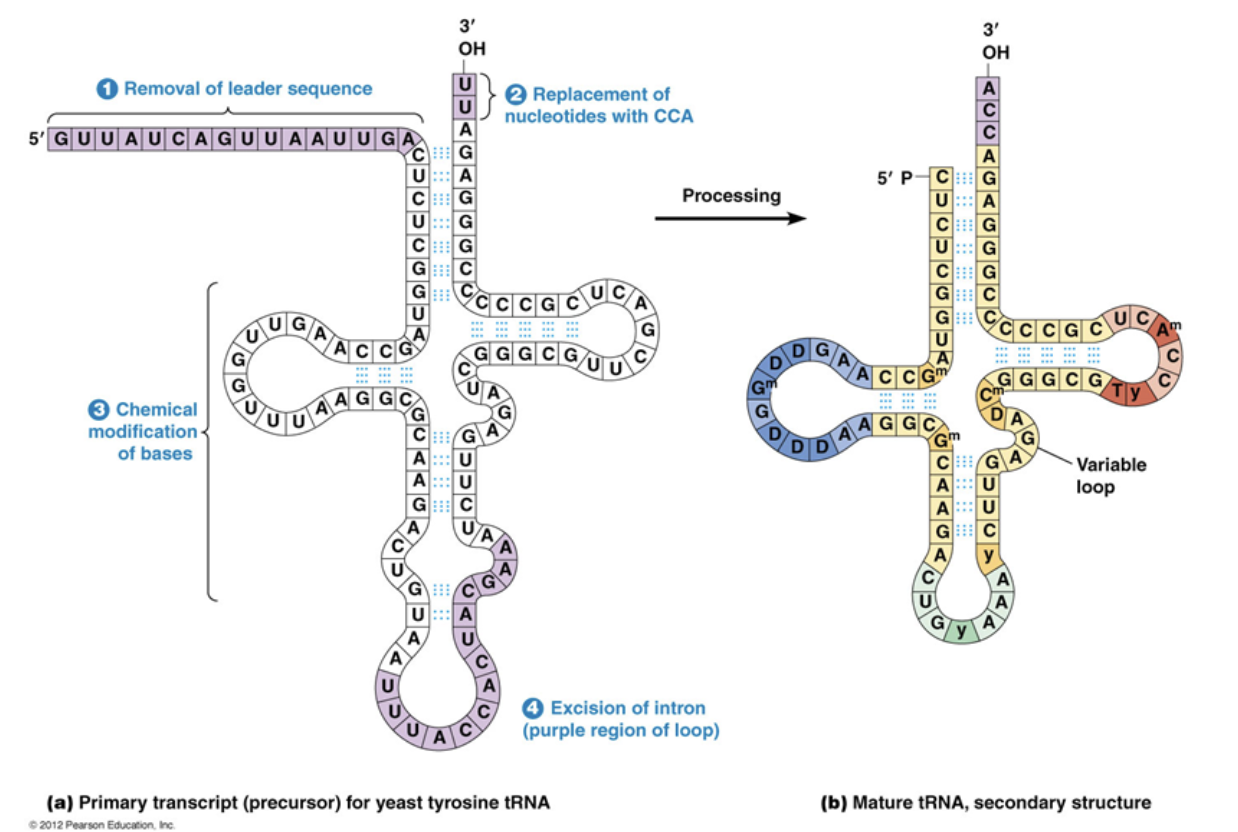
Which four things occur during tRNA processing?
The leading sequence is removed at the 5’ end, one nucleotide at a time.
Nucleotides are removed and then replaced by 2 new nucleotides at the 3’ end
Bases making up the secondary structure are chemically modified and made into nucleotides specific to tRNA synthesis
Introns are spliced out
Ribosomal RNA (rRNA)
Synthesized by RNA polymerase I and III
Form ribosomes and therefore work as a catalyst to protein synthesis
The ribosome has 4 RNAs which catalyze translation
3 of them are transcribed as a single unit. After they’re transcribed, phosphodiester bonds break up via cleavage to form 3 functional units
Micro RNA (miRNA)
Synthesized by RNA polymerase II and III
Act as regulators for protein synthesis and translation
They are complementary to mRNA and can either trigger mRNA degradation or translation inhibition
When miRNA is partly bound to mRNA in a certain way, it blocks RNA polymerase from being able to read the code, leading to translation inhibition.
If miRNA is fully bound and complementary to mRNA, it acts as a recognition site for nucleases to cleave the mRNA so that it can no longer be translated, leading to mRNA degradation
What three main components are required for translation?
The genetic code, made of mRNA
The translator, made of tRNA. Contains a complementary portion to the mRNA so it can read the RNA language
The enzyme that brings them together, the ribosome made of rRNA and proteins
Translation is coded for by mRNA and read by tRNA. This process is catalyzed by rRNA and regulated by miRNA.
Describe initiation in translation
The first step of translation where the code, translator, and ribosome all come together
The small ribosomal subunit binds to mRNA with the help of a g-protein called IF-2
The ribosome has three different slots in which the tRNAs can sit
Initiation factors bind to the A and E sites, leaving the P site the only one open for the first tRNA to bind.
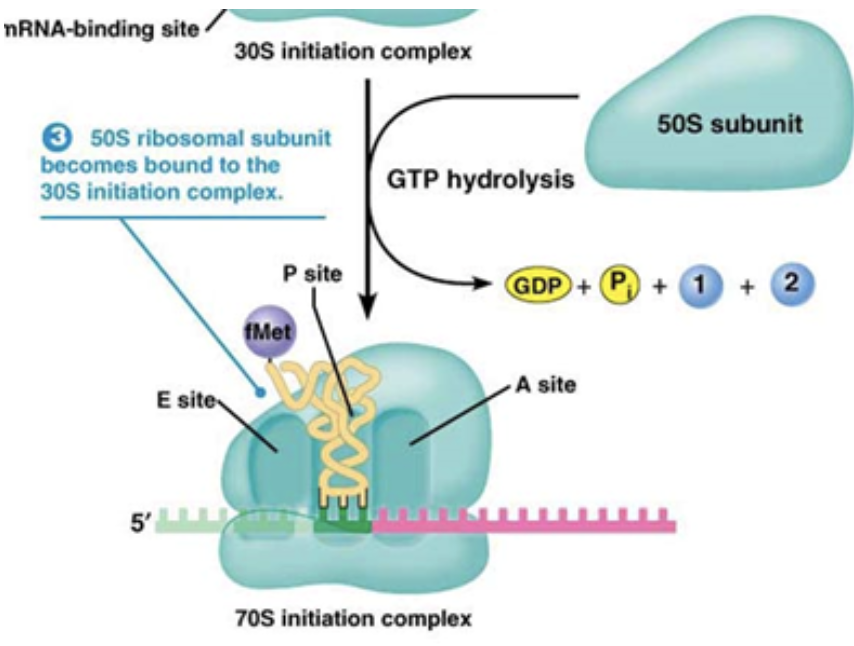
What are the three sites of the ribosome?
The right site, Amino-acyl (A) site, where new RNAs enter along with their amino acids
The middle site, peptidyl (P) site, where the peptide chain sits as it grows
The left site, exit (E) site, where tRNAs exit the ribosome without their amino acids
The first tRNA in initiation always binds to the P site, while all subsequent tRNAs will bind to the A site first
Describe elongation in translation
A new tRNA, with its amino acid bound, enters the A site depending on its complementary anti-codon and the codon of the mRNA. This is assisted by EF-Tu
A peptide bond is formed when the new amino acid is added to the old amino acid and is transferred from the A site to the P site. This is a condensation reaction. The newest amino acid will always be at the bottom.
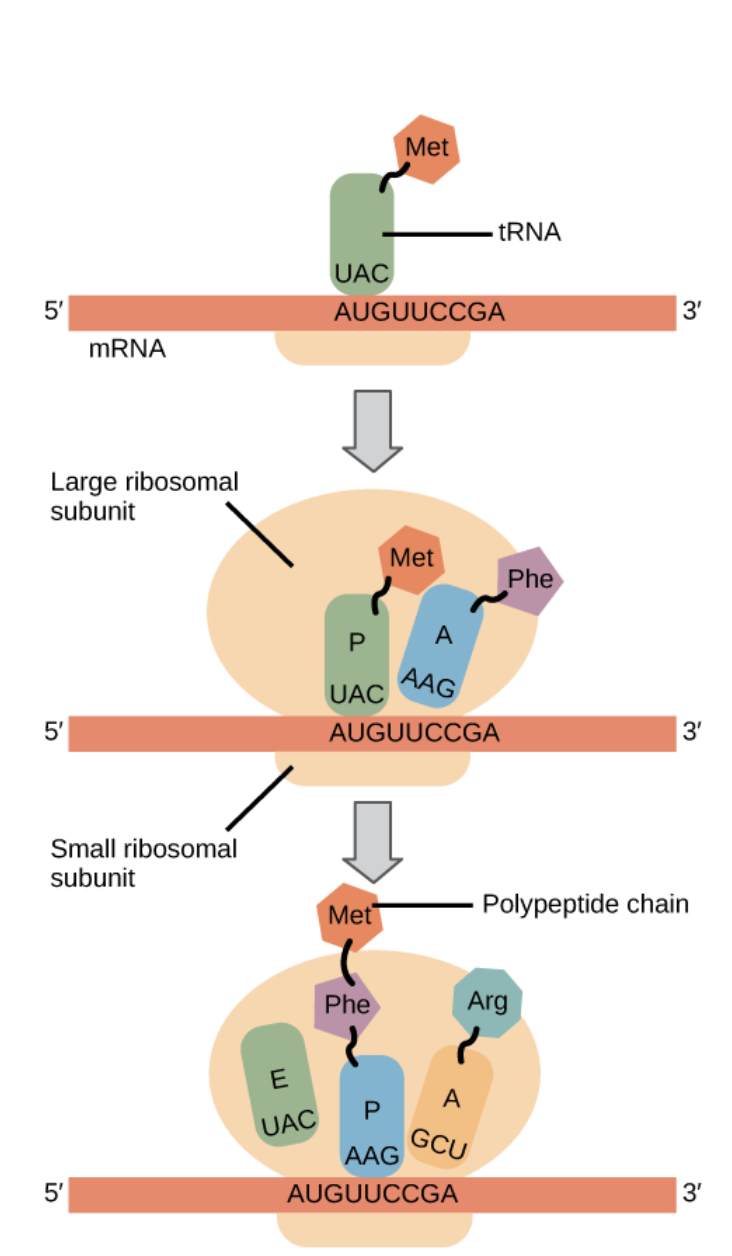
Describe termination in translation
Occurs when the mRNA reaches it’s stop codon. The stop codon is bound to a g-protein called the release factor
Once the ribosome reaches the stop codon, a protein is added rather than a tRNA. This protein causes the ribosome, mRNA and tRNA structure to fall apart via allostery
When the final condensation reaction occurs, water is just made and amino acids are no longer added
Hydrophobic interactions cause the secondary structure of a protein to be formed as soon as the protein starts to emerge from the ribosome
IF-2
Helps the small ribosomal subunit bind to mRNA in initiation of translation
When the large ribosomal subunit binds next overtop the mRNA code, it causes initiation factors to be lost. GTP is hydrolyzed to GDP, causing allostery
Initiation factors bind to the A and E sites, leaving the P site the only one open for the first tRNA to bind.
EF-Tu
Helps a new tRNA enter the A site in elongation of translation
EF-Tu uses a guess and check method. tRNAs are attached to EF-Tu. If the mRNA and the tRNA are a match, then EF-Tu has its GTP hydrolyzed to GDP and it leaves due to poor binding affinity.
If they are not a match, tRNA attached to EF-Tu leaves, and tries again on another ribosome
EF-G
Helps stabilize the small ribosomal subunit as it translocates during elongation of translation
It hydrolyzes GTP into GDP. This hydrolyzation pulls the small ribosomal subunit up to meet the large subunit. It goes in the A site and then exits. The complex is now stable again and has moved over 3 nucleotides.
Release factor
Bound to the STOP codon and reads it once it is reached by tRNA. Assists termination in translation
It looks like a tRNA, but has no amino acid bound to it.
A g-protein. When the release factor hydrolyzes GTP into GDP, it provides the energy needed to break the complex of mRNA, tRNA, and the ribosome
Why can’t proteins that will eventually be membrane bound or secreted be translated within the cytoplasm?
Because these proteins are largely hydrophobic
Because of this, it is necessary for proteins to be membrane proteins from the time they are translated. This means that they have to be a part of the endomembrane system
Membrane bound/secreted proteins start translation on a free ribosome and end it within the ER. This is indicated by a code in the mRNA
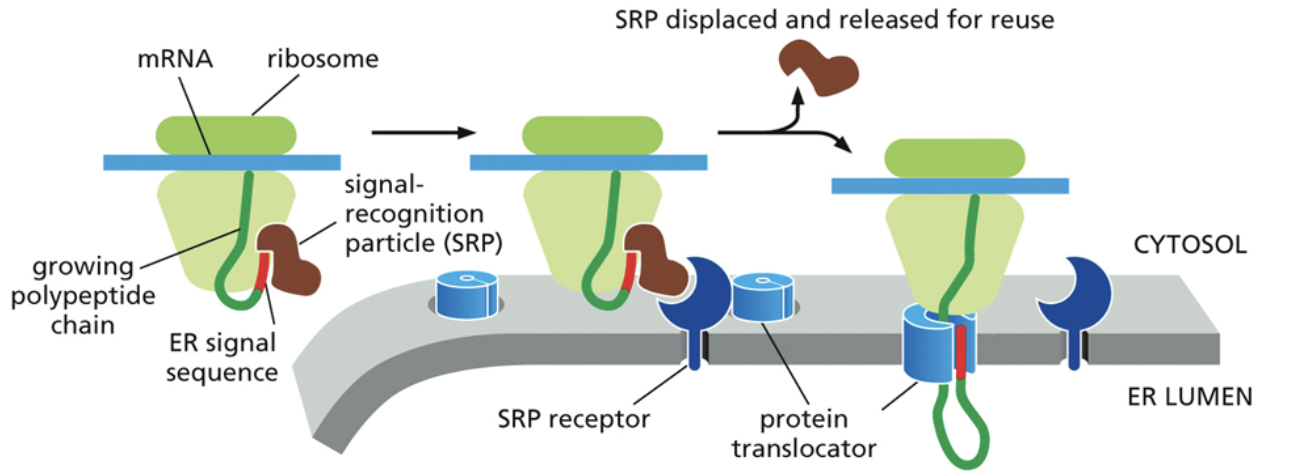
How are proteins transported and translated to the ER?
Transport to the ER begins with the recognition of a signal sequence by the SRP. Within the free ribosome, SRP first binds to the end of a protein that is currently being synthesized.
It then binds to the SRP receptor (a g-protein) at the same time, located atop the ER. The SRP receptor is a part of the translocon
This binding brings down the ribosome, protein, and mRNA where the protein will continue translation directly through the pore and into the ER.
The signal proteins are then cleaved after translation so it doesn’t just get brought back to the ER
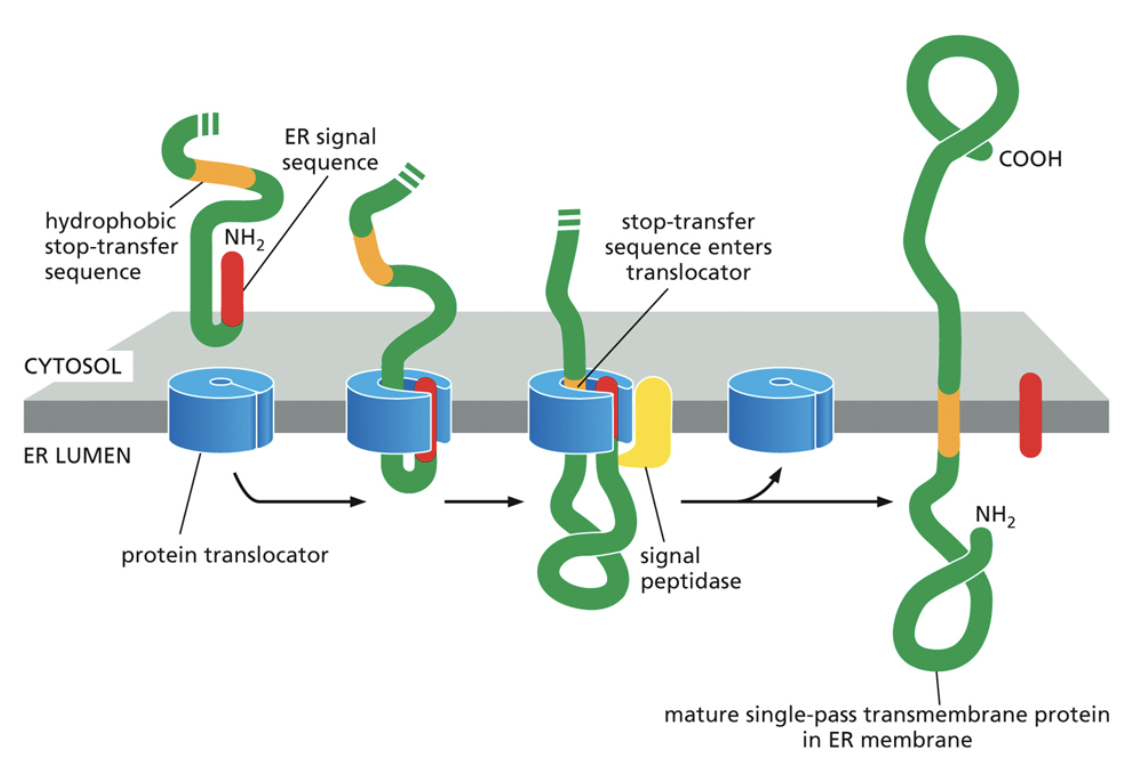
Why is the STOP codon of a transmembrane protein located inside of the ER membrane?
This component is very hydrophobic and cannot end up in the cytoplasm or the ER lumen
When the stop transfer sequence ends up in the middle of the translocator, it will open up and the protein will float out into the membrane to continue translation
The oldest part of the protein ends up in the ER lumen, the stop sequence ends up within the membrane of the ER, and the rest ends up in the cytoplasm
What are the functions of the Endoplasmic Reticulum (ER)?
The ER performs quality control on the function for proteins
The ER has a mechanism that ensures proteins are properly folded before being sent to the Golgi
The ER is the site of translation for secreted and transmembrane proteins
How does the ER ensure that proteins are properly folded?
This process begins with the addition of sugars on the nitrogens in proteins. Each protein gets the same sugar because they’re being checked by the same mechanism. This is called N-linked glycosylation.
Once the sugars are on the protein, the sugars get bound to a recognition protein, which checks for proper folding.
The recognition protein is called calnexin. It is bound to the ER membrane and a binding site for the sugars. It looks for hydrophobic things that do not belong. If calnexin finds something hydrophobic, it will send the protein back by cleaving its sugar
What happens to proteins that cannot be properly folded within the ER?
Proteins that cannot be folded act as sensors to signal problems within the cell.
They bind to transmembrane proteins in the ER. These transmembrane proteins will activate if there are too many misfolded proteins bound to them
Once it activates, it may create translation inhibitors so the ER does not continue to get overworked
Misfolded proteins can also activate chaperone genes that increase the protein folding capacity of the ER
What are the functions of the Golgi Apparatus?
To sort proteins so that they can be delivered to their final destinations
They do this by adding sugars individually to oxygens on proteins, one sugar at a time. The combination of sugars can be read to determine the protein's final destination. This is called O-linked glycosylation
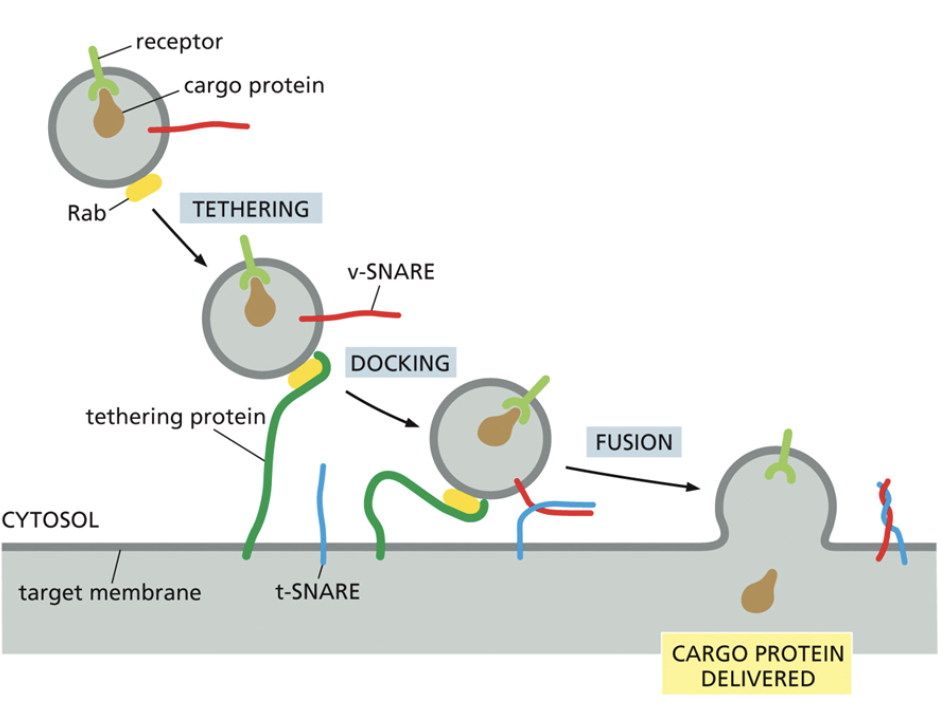
Sugar combinations are recognized by cargo receptors, which begins vesicle formation
Adaptin
A g-protein bound to receptors on vesicles that read sugars added during O-linked glycosylation
When they are bound to GTP, they’re curved, allowing the vesicle to have a circular structure.
Adaptin is bound to coat proteins. There are different kinds of coat proteins depending on the final destination of the cargo the vesicle is carrying.
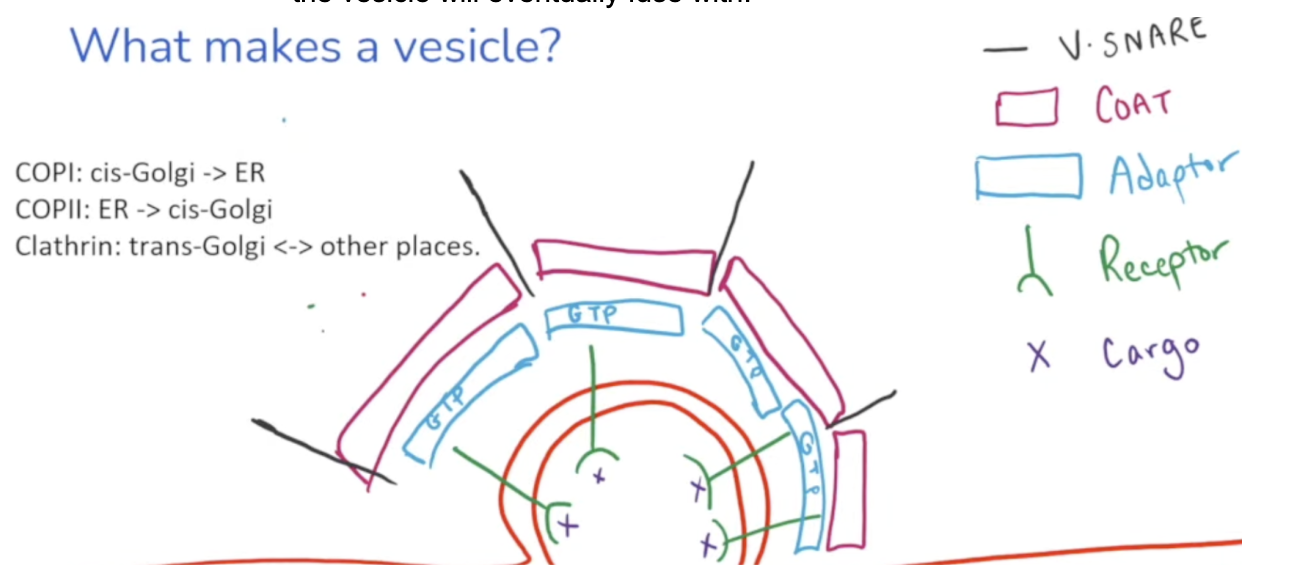
COP I
A coat protein on a vesicle that carries cargo from the cis Golgi → ER
COP II
A coat protein on a vesicle that carries cargo from the ER → cis-Golgi
Clathrin
A coat protein on a vesicle that carries cargo from the trans golgi ←> other places
V-snare
Additional proteins located outside the vesicle
Have a complementary T-snare that is bound to the membrane the vesicle will eventually fuse with
Snare proteins recognize final destinations and tether proteins to pull them closer for fusion. GTP is hydrolyzed to linearize the vesicle for fusion
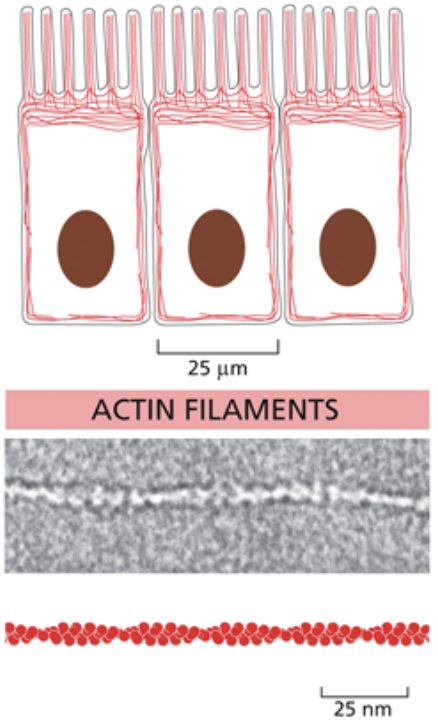
Actin/micro filaments
The smallest cytoskeletal molecule
They’re made of filaments that coil around each other to form rope like structures, but they don’t repeatedly coil like intermediate filaments
Also unlike intermediate filaments, they are directional and asymmetrical
Microfilaments are made of the monomer g-actin. When g-actin binds to ATP, it is activated due to allostery.
Actin can take on a variety of shapes depending on its function in specific cells. Can run parallel or antiparallel
Enable cell crawling and muscle contraction
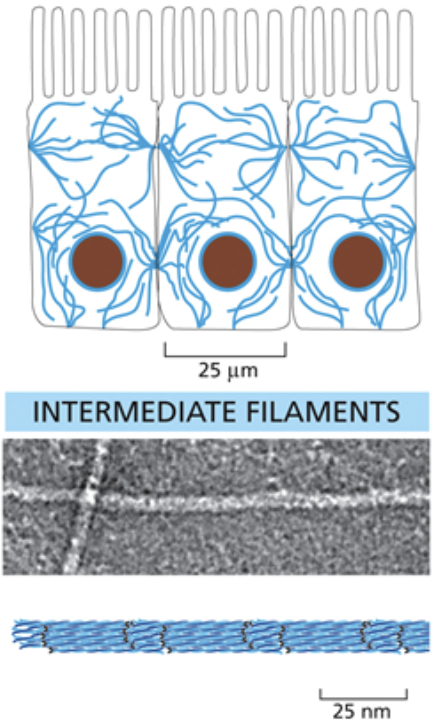
Intermediate filaments
Midsized cytoskeletal molecule
They are structured as symmetrical dimers; Long protein strands coil around each other, then those coils coil around each other. This makes it so it can snap without fully breaking
Intermediate filaments are intermediately spaced within the entire cell and don’t have a clear formation
These strands run antiparallel and do not have directionality
They are made of a few different types of monomers
They are usually connected to the membranes of cells to relieve mechanical stress and organize the cell
Intermediate filaments link to the rest of the cytoplasm and anchor organelle using plakins
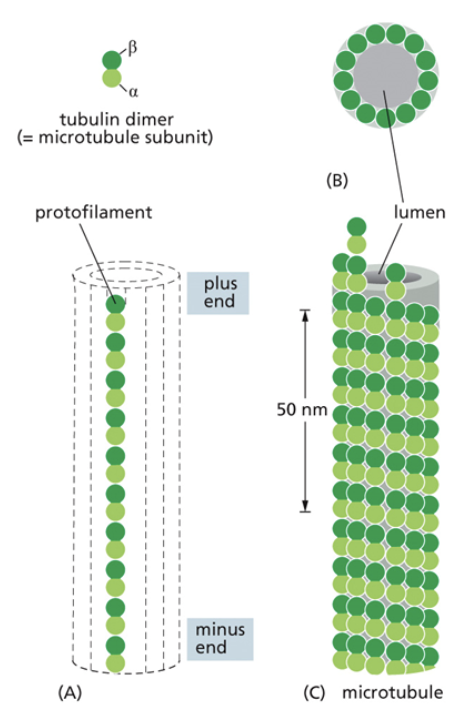
Microtubules
The largest cytoskeletal molecule
Act as “roads” for motor proteins
They stem from the centromere
They are made of dimers that form filaments, and then tubes. Dimers are made of two subunits called alpha and beta tubulins. Dimers are asymmetrical
Tubulin molecules are g-proteins. They are active when they are bound to GTP. They are inactive and likely to collapse when bound to GDP
There is a + and - end of filaments, but this has nothing to do with the molecules charge. The + end is the beta end, the - end is the alpha end.
They are organized at their negative end by microtubule organizing centers (MTOCs)
Dynamic instability
The rapid and frequent growth and shrinking of microtubules
Cytoskeletal molecules can grow and shrink as needed
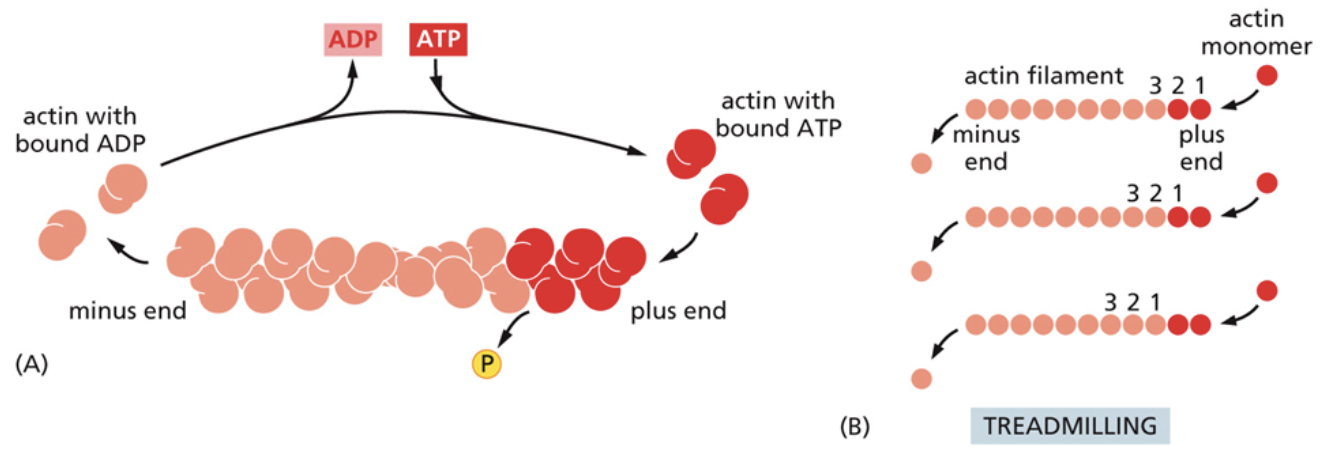
Treadmilling
A process used by microtubules and microfilaments to recycle their subunits
Subunits are added from one end and subtracted from the other. This moves all monomers to the left
This helps control the length of microtubules and microfilaments by determining how quickly monomers are added and subtracted.
For example, if monomers are being subtracted quicker than they are added, the filament shrinks
Catastrophe
Microtubules can shrink rapidly from the + end by peeling back filaments like a banana peel
This is caused by ATP hydrolysis
This occurs during mitosis, when the cytoskeleton moves chromosomes
What determines how cytoskeletal molecules shrink and grow?
Proteins!
This regulation allows the cell to grow in specific directions, such as when the cell is crawling
Cell crawling
The process of a cell moving toward or away from stimuli, done by the cytoskeleton
Requires cell protrusion, cell contraction, and moving molecules within the cell
Protrusion is the cells cytoskeleton growing at the front in the direction the cell is trying to move
Contraction is the cell pulling up on the back of the cell so it moves forward
Molecules in the cell are moved due to cell crawling when they are put in vesicles and delivered to the front
Kinesin
Move cargo toward the + end of the microtubule
Dynein
Move cargo toward the - end of the microtubule
What are parts of both kinesins and dyneins?
Globular head- The part that connects to the microtubule. Looks like feet
Coil coil region- Absorbs tension caused by the steps they take
Binding cargo region- Where vesicles, organelle, or other cargo bind to be carried to their destinations
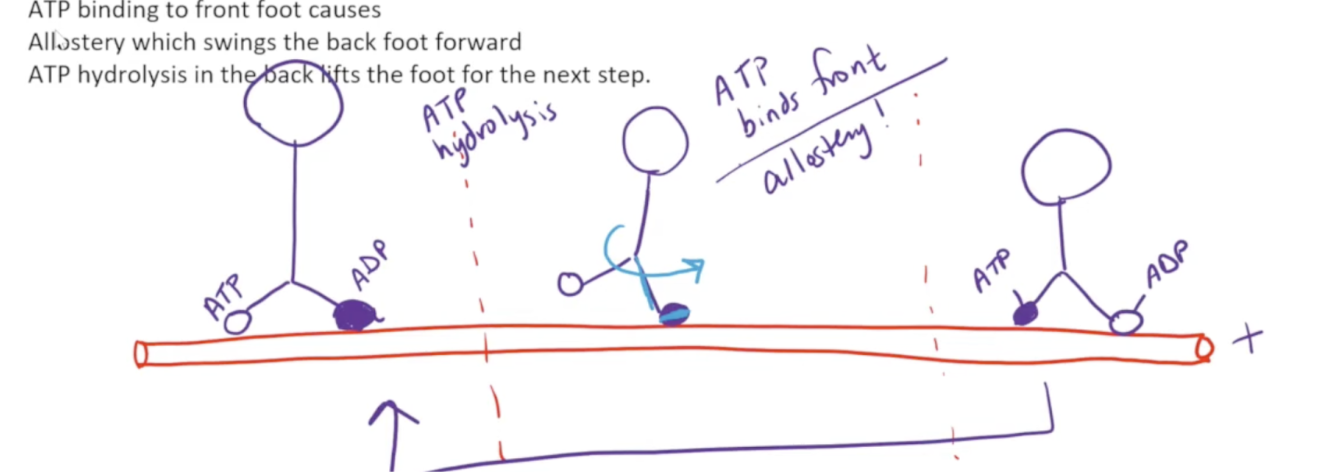
What are the three steps kinesins and dyneins perform to move across the microtubule?
When ATP is bound to the globular head, it is harder to remove. To begin with, the front foot of the globular head is bound to ADP whilst the back foot is bound to ATP. ATP hydrolysis occurs, which causes the back foot to be removed from the microtubule
ATP then binds to the front foot. This causes a linker region to swing forward and zip along the front foot. This swings the molecule so the back foot ends up in the front
ATP hydrolysis occurs, and the back foot is once again released, leading to the steps repeating
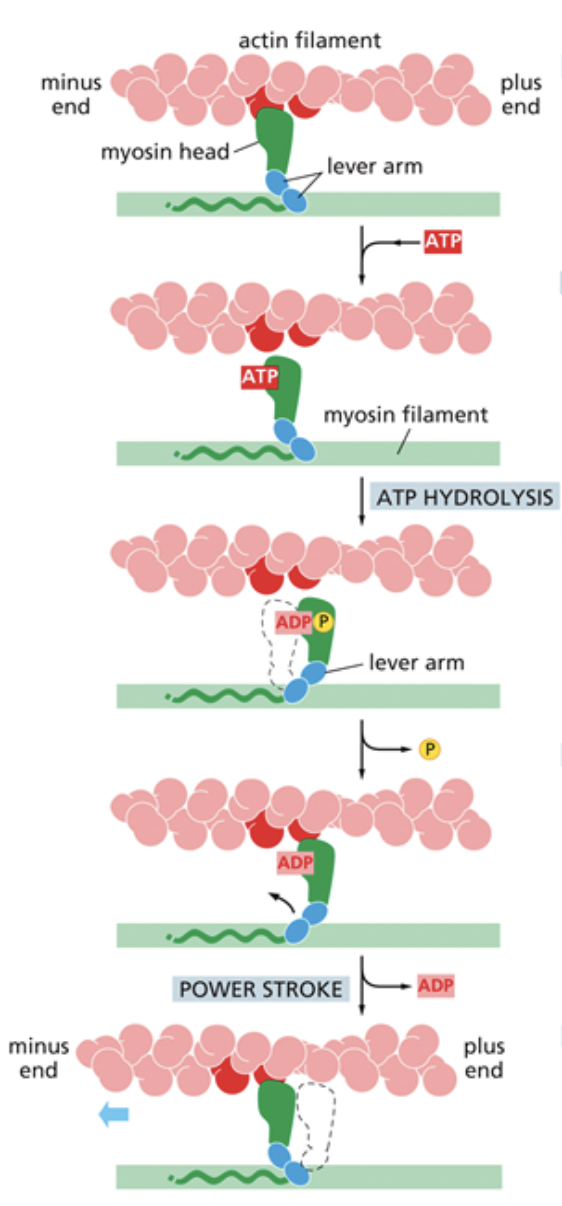
Myosin
Move cargo along microfilaments with a domain that binds to the MF. The way it takes steps is different from kinesins.
They can contract the cell by moving microfilaments closer together. It does this by pulling on either side of the cell membrane
What are the steps myosins take to contract and move cargo?
ATP binds to the myosin head and causes an allosteric change that makes the myosin release from the microfilament
ATP hydrolysis occurs, which pulls the myosin back into the cocked position, like a sling shot
The phosphate from ATP hydrolysis is released, which causes the tension from the cocked position to be released
Myosin swings because of the release of tension and latches back onto the microfilament. Called the power stroke. ADP is released, causing the myosin to go back to the position it was in before step one and allowing ATP to bind once again.