Exam 1
1/77
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
78 Terms
What are the stages of the cell cycle and what are their names?
M = mitosis
G1 = gap phase
S = DNA synthesis
G2 = second gap phase
G0 = phase where cell leaves the cell cycle
What are the 3 cell types (cell cycles in vivo) that are distinguished based on their capacity to grow and divide and what are some examples?
Highly specialized cells that lack the ability to divide, once they differentiate they remain in that state until they die. ex. nerve and muscle cells or RBCs
Cells that normally don’t divide but can be induced to divide given an appropriate stimulus. Ex. liver cells and lymphocytes.
Cells that normally posses a relatively high level of mitotic activity. Ex. hematopoietic stem cells.
What is the central dogma of biology?
DNA undergoes transcription to become RNA which undergoes translation to become a protein.
What is different about prokaryotic and eukaryotic chromosomes
Prokaryotic = single circular chromosome plasmid (no end to it) 5.6 × 106 bp that have 4,300 protein-encoding genes.
Eukaryotes = linear, 3 × 109 bp DNA per cell. We have 20,000-25,000 protein-coding genes.
What do unicellular organisms to make copies of themselves?
Binary Fission
How many chromosomes do we have? How many do we get from our parents? How many autosomes versus sex chromosomes do we have?
We have 46 chromosomes, 23 from mother and 23 from father. If an organism has 2 sets it’s diploid. We have 22 sets of autosomes, and one set of sex chromosomes. (X or Y)
What is included during interphase and mitosis of the cell cycle? What is the point in G1 where cells are nondividing?
Interphase includes = S phase (DNA is synthesized), and G1 and G2
G0 - point in G1 where cells aren’t dividing but in a metabolically active state
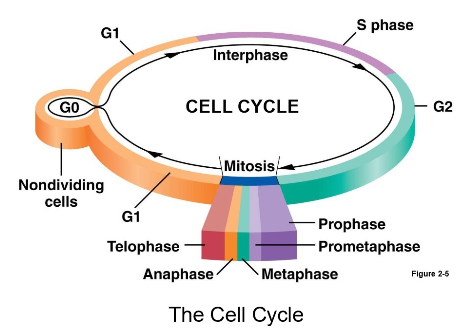
What are the 3 checkpoints in the cell cycle and what is being evaluated then?
G1/S checkpoint = monitors size cell has achieved (big enough to divide?), evaluates condition of DNA (DNA integrity)
G2/M checkpoint = monitors if DNA replication is incomplete (pause cell if there is damage) monitors damaged DNA
M checkpoint = monitors successful formation of spindle fibers system and attachment to kinetochores
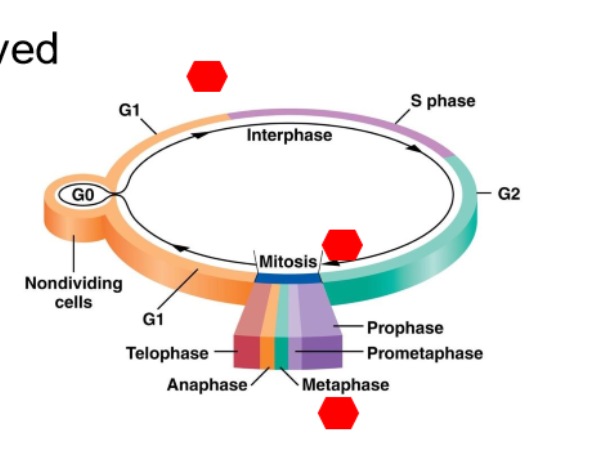
What is the result of cell cycle in a diploid cell?
Two daughter cells with a full diploid complement of chromosomes and are genetically identical to the parent cell
Quiz:
Immediately after a cell is made by its parent cell undergoing M phase, that cell is in what phase?
A. G1 (part of interphase)
B. S (part of interphase)
C. G2 (part of interphase)
D. M (mitosis)
A. G1 (part of interphase). Immediately after one cells divides into two cells, those two cells will be in G1 phase (see slide 13).
A cell that leaves the cell cycle is:
A. dying or dead.
B. in G0.
C. in M.
D. in a gap phase (G1 or G2).
B. in G0. See slide 13
ls in G0 are resting.
True. Because cells are not actively in the cell cycle, they don't do anything else.
False. While they're not progressing through the cell cycle, they're metabolically active, doing the things the cell needs to do.
False. While they're not progressing through the cell cycle, they're metabolically active, doing the things the cell needs to do.
A differentiated cell (not a stem cell, and it's not planning to divide again so has left the cell cycle) will do what it needs to do (muscle cells will contract and relax, nerve cells will send and receive cellular messages, etc.).
What is the checkpoint that confirms that DNA replication is complete and error-free, and no DNA damage exists?
A. G1/S checkpoint
B. G2/M checkpoint
C. S/G2 checkpoint
D. M checkpoint
E. G0 checkpoint
B. G2/M checkpoint. There are only three major checkpoints in the cell cycle. See slide 14 for the one that occurs after DNA replication (S phase). This checkpoint needs to give the "all clear" signal before the cell enters M phase, so the two daughter cells get full, error-free copies of the parent's DNA!
What is each phase of the cell cycle known for (G0, G1, G2, M, S)
G0 Cells are not dividing but in a metabolically active state
G1 Cell growth and normal metabolism, prep for DNA replication
G2 Further cell growth, synthesis of proteins needed for mitosis, prep for cell division
M Mitosis, division of nucleus and cytoplasm
S Synthesis, DNA replication
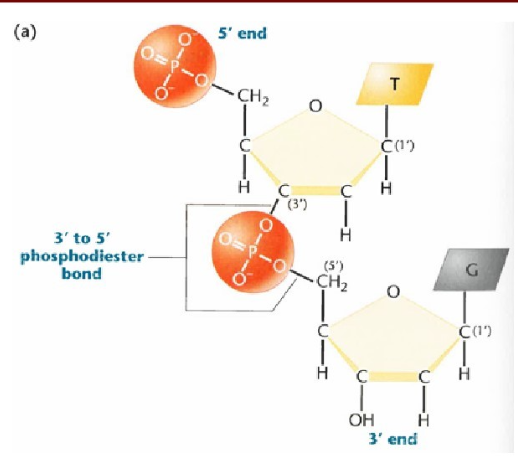
What order is DNA made? and What differentiates those ends?
DNA is made 5’ end to 3’ end.
The 5’ end has a phosphate group attached to the 5th carbon of the sugar
The 3’ end has a free hydroxyl group attached to the 3rd carbon of the sugar
What does the Watson-Crick model suggest about the what functions as the template for synthesis of new DNA strands?
The Watson-Crick model suggest that the two strands each serve as a template for synthesis of new DNA strands.
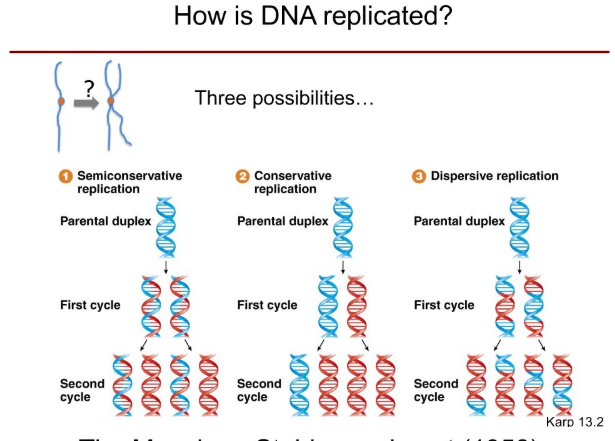
What are the 3 possibilities of DNA replication?
Semiconservative replication: Parental DNA strands are conserved but new DNA strands are added onto the parental ones.
Conservative replication: Parental duplex retains it’s sequence when a new piece of DNA is made, both of the strands of one are new.
Dispersive replication: The two duplicated daughter strands would have some chunks of new DNA and some parental DNA.
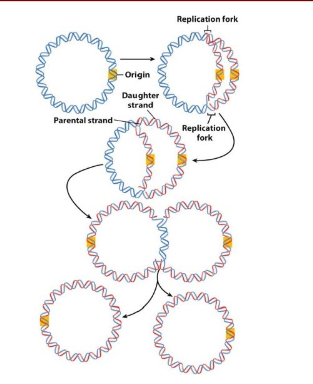
How do bacteria (prokaryotic) replicate their chromosome?
Start at Origin of replication
Replicate DNA bi-directionally at replication forks (where strands seperate)
terminate replcation when the replication forks meet at the opposite point on the circular chromosome
seperate circles
cell replication
Where does replication start in eukaryotes?
Eukaryotes replicate their genome in small sections called replicons which are replication bubbles.
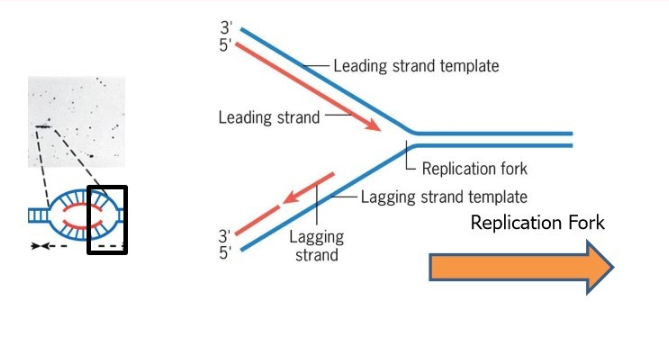
How is DNA being read and how is it being created? What makes a leading versus lagging strand
DNA is read from the 3’ to 5’ while it’s created from 5’ to 3’ end.
leading strand is the one being created on the 3’ to 5’ template strand
lagging strand is the one being created on the 5’ to 3’ template strand
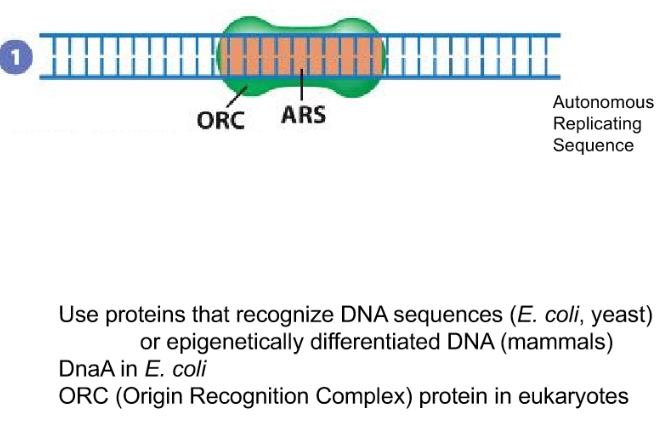
What proteins are used to recognize the replication start site in DNA sequences for prokaryotes versus Eukaryotes?
Prokaryotic: DnaA in E.coli
Eukaryotic: ORC (Origin Recognition complex) will recognize the ARS (autonomous replicating sequence)
What are the two major nucleotide requirements for replication? What makes a good version of these two things?
A template. A good template needs to be single-stranded, non circular (bc no primer),
A primer - second strand of nucleotides (DNA or RNA) that has to be laid down before the polymerase can start. A good primer would have a section of DNA that has a gap in it with an available 3’
What proteins are used to unwound a double helix to make a template for replication for prokaryotes versus eukaryotes?
Use a helicase to unwind the dsDNA and make it single stranded. And use a topoisomerase to relieve the tension created by the helicase
Prokaryotic: DnaB as a helicase and gyrase as a topoisomerase
Eukaryotic: Mcm2-Mcm7 six protein complex in eukaryotes as a helicase and Topoisomerase I/II as a topoisomerase.
What tells a helicase to start in prokaryotes and eukaryotes?
Prokaryotic: DnaC load the helicases onto the DnaB helicase.
Eukaryotic: Cdc6 ATPase, Cdt1 loads 2 helicase complexes onto the arc to form a pre-replication complex (pre-RC).
Cyclin dependent kinase (Cdk) and other cell cycle regulated proteins initiate replication (also prevents reloading of helicases during the same cell cycle)
How does the single stranded DNA remain open until DNA replication occurs in prokaryotes and eukaryotes?
Attach single stranded DNA binding protein to the ssDNA
Prokaryotic: Single-stranded DNA binding proteins (SSB)
Eukaryotic: RPA (Replication protein A) in eukaryotes
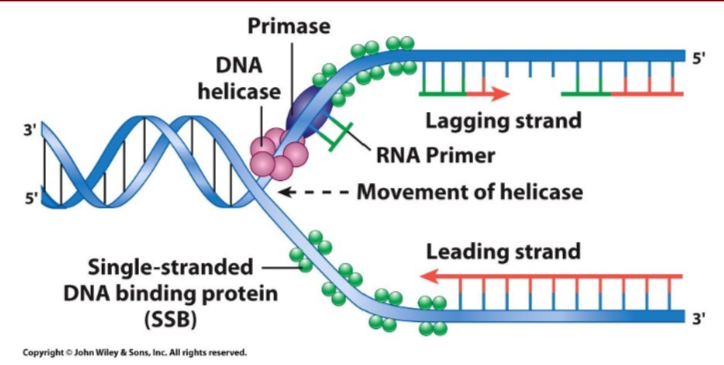
How is the primer added to the template DNA in prokaryotes and Eukaryotes? What does the helicase and primase form along the lagging strand?
Primase synthesizes short RNA primer (one on leading strand because it’s continous, multiple on lagging strand because it’s discontinious)
Prokaryotic: Primase
Eukaryotic: subunit of pol a (alpha) in eukaryotes. Pol a creates an RNA primer and then makes a short DNA chain.
A primosome is formed from the helicase and primase along the lagging strand.
If you start with a cell that has one chromosome, how many chromosomes (total, in the resulting cells) will you have after two cell cycles?
A. 2
B. 4
C. 8
4. A cell with one chromosome goes through one cell cycle, resulting in two cells (two chromosomes). After the second cell cycle, those two cells will have each divided, making four cells total. Therefore you'll have four chromosomes after two cell cycles.
If you label a single parental chromosome and not newly synthesized DNA strands, how many chromosomes will be labeled after one cell cycle?
A. 0
B. 1
C. 2
D. 4
E. 8
C. 2; In the parental chromosome, there are two DNA strands. Because of semi-discontinuous replication, one of those two parent strands will be in each daughter chromosome. So the answer is two chromosomes will be labeled after one cell cycle!
If you label a parental chromosome and not newly synthesized DNA strands, how many DNA strands will be labeled after two cell cycles?
A. 0
B. 1
C. 2
D. 4
E. 8
C. 2;The same two parent strands that were labeled will remain labeled, but they'll be in two different chromosomes in two cells after two cell cycles.
What protein complex identifies and binds the replication start site in eukaryotes
(including us)?
A. SSB
B. ORC
C. DnaA
D. helicase
B. ORC; In eukaryotes, the ORC (origin recognition complex) identifies the start site. In prokaryotes, the start site is recognized by DnaA.
What protein complex unwinds dsDNA in eukaryotes
(including us)?
A. SSB
B. ORC
C. DnaA
D. helicase
D. helicase; Helicase is the protein complex in us and other eukaryotes that rips apart the double-stranded DNA to make a template for replication.
With an open template and a primer loaded in place, how does replication occur in prokaryotes and eukaryotes? (what does the complex contain?)
To make a replisome, a complex containing a polymerase holoenzyme, helicases, topoisomerases, SSBs and other associated proteins.
Prokaryotic: Pol III
Eukaryotic: Pol ϵ for the leading strand and Pol δ for the lagging strand. They replace pol alpha on the template strand.
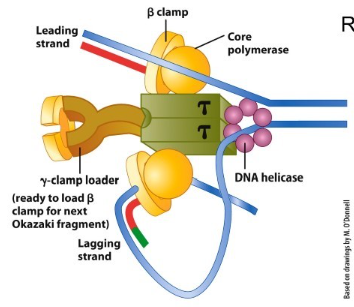
What are the two requirements that a polymerase needs to do to stay on? How does polymerase stay on and move along the DNA in prokaryotic and eukaryotic?
The polymerase needs to 1. remain associate with the template over long stretches and 2. be attached loosely enough to move one nucleotide to the next
The polymerase binds to a sliding clamp.
Prokaryotic: Beta clamp
Eukaryotic: PCNA (proliferating cell nuclear antigen)
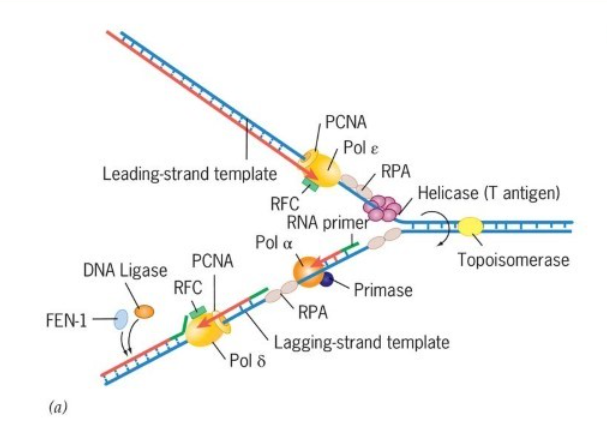
How does the sliding clamp get loaded onto DNA at the replication in Prokaryotes and Eukaryotes?
A clamp loaded complex that loads each clamp onto DNA, keeps polymerases on both DNA strands in the replisome, and bind to helicase to keep the complex at the replication fork.
Prokaryotic: Gamma (𝛾) and Tau (𝜏) apart of the pol III complex
Eukaryotic: Replication factor C (RFC)
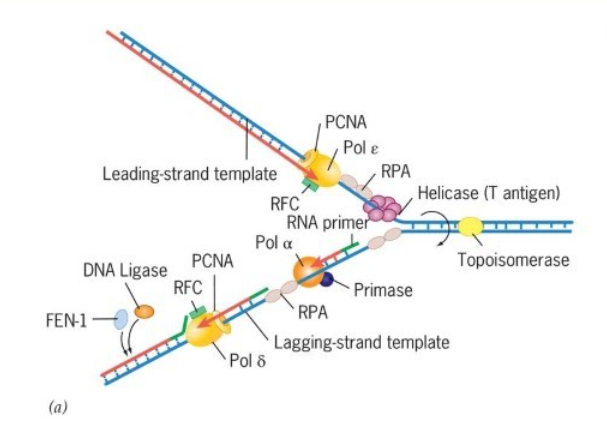
What is the trombone model of DNA replication?
describes how the lagging strand of DNA loops out, like a trombone's slide, to allow both leading and lagging strands to be synthesized simultaneously at the replication fork by coupled enzymes, coordinating the continuous leading strand with the discontinuous, fragment-by-fragment (Okazaki fragments) synthesis of the lagging strand, with the loop growing and then releasing as the polymerase hops to the next site.
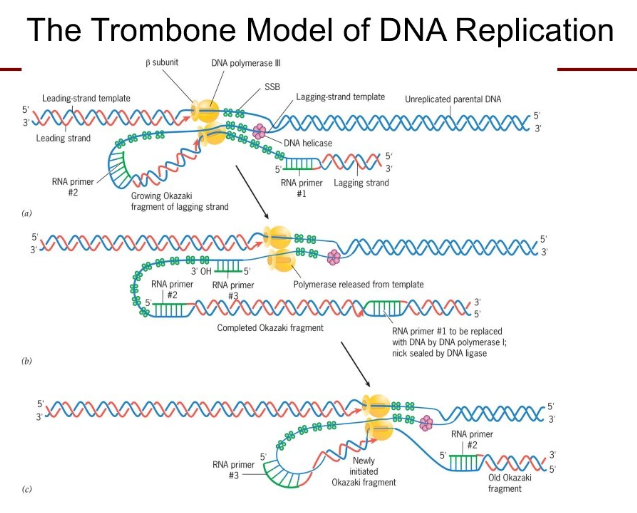
With millions of nucleotides to replicate so fast, how does polymerase do it so well? (how does the proofreading function work?)
Improper geometry: Typically the mismatched nucleotide doesn’t fit in the binding pocket of the polymerase. But if it is incorporate..
The polymerase has proofreading functions too. 3’ → 5’ exonuclease activity. the misincorporated nucleotide has an increased tendency to separate from the template and “fray.”
the polymerase stalls
the new DNA strand end is rotate into the exonuclease pocket of the enzyme and it’s removed (3’ to 5’)
Then the new DNA strand is shifted back into the polymerization pocket of the enzyme
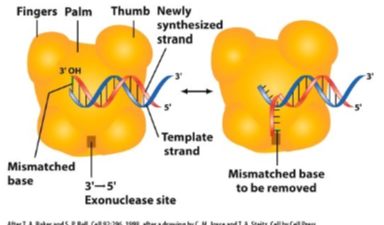
What are the 3 reasons as to how DNA is replicated so accurately?
The polymerases are great at accurately selecting nucleotides
The polymerases proofread, remove incorrect nucleotides, and replace them with correct nucleotides
Post replicative mismatch repair system fixes most remaining errors.
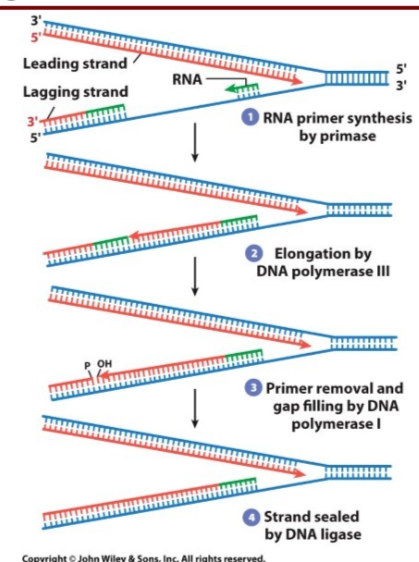
How are the ends taken care of in Okazaki fragments in prokaryotes and eukaryotes?
Prokaryotic: after DNA pol III drops off lagging strand, DNA pol I enters and removes RNA primers (5’ → 3’) exonuclease activity, polymerizes new DNA, then proofreads it using exonuclease activity
Eukaryotic: pol δ displaces the RNA primer and FEN-1 RNaseH endonuclease cuts the RNA primer off
How is the gap of unreplicated DNA left at the extreme 5’ end of the DNA synthesized by lagging strand replication solved? (what are it’s 2 main components?)
The use of Telomeres and telomerase.
TERT is a reverse transcriptase
TER is a RNA subunit with a sequence complementary to the telomere repeat sequence. TER acts as a template for the reiterative addition of G-rich telomeric repeats to the chromosome 3’ end.
How do Telomeres and telomerase function to sovle the end replication problem?
Once the 3’ end of the DNA is extended by telomerase, a new RNA primer can be synthesized by primase close to the chromosome terminus.
The primer will be extended by DNA polymerase just like in the body of the chromosome
Although there will still be a small gap on the chromosome end (3’ overhand) when the last primer is removed, the reiterative nature of telomere repeats ensures that no essential genetic information will be lose.
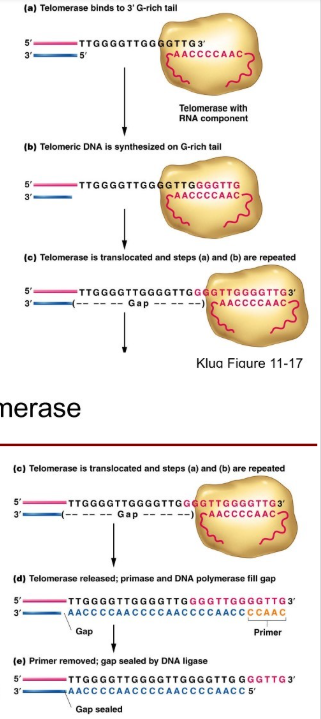
How do Telomeres protect their ends? (including how some eukaryotes do)
They protect their ends with proteins and in some eukaryotes a loop.
Telomere-specific proteins distinguish the chromosome terminus from a double strand break (damaged DNA) they protect the telomeres ends from attack by nucleases
What protein binds ssDNA to keep it open for replication?
a. SSB
b. ORC
c. DnaA
d. helicase
Correct answer: a. SSB; Singled-stranded DNA binding proteins (SSB) binds to single-stranded DNA to keep it open for the replication complex to do its job of DNA replication.
DNA polymerases not only create a new strand of DNA from a single-stranded template, they also:
a. unwind the double-stranded DNA into two single strands.
b. proofread their work, using a 3'-to-5' exonuclease activity to remove mis-paired nucleotides to remove ~99/100 potential mutations.
c. create RNA from the DNA template.
d. create protein from the DNA template.
e. load themselves onto the DNA.
b. proofread their work, using a 3'-to-5' exonuclease activity to remove mis-paired nucleotides to remove ~99/100 potential mutations.
What protein seals nicks in the newly synthesized strand of DNA?
a. DNA pol III
b. DNA pol I
c. FEN-1
d. DNA ligase
d. DNA ligase; DNA polymerases can't do that last step of joining new segments of DNA together. FEN-1 RNAseH is an endonuclease that cuts the RNA primer off in eukaryotes before the DNA ligase seals that final phosphodiester bond between nucleotides in the newly synthesized DNA strand.
Telomerase is an enzyme with both protein and RNA components! What is TERT?
a. the reverse transcriptase (protein part of the enzyme)
b. the RNA part of the enzyme
c. the T-loop made by telomerase
d. a topoisomerase
a. the reverse transcriptase (protein part of the enzyme); TER is the RNA subunit, and TERT (think of it as TER Transcriptase) is the protein/enzymatic part.
What is the definition of a mutation? What are the example of spontaneous mutations? What can cause induced mutations?
a Mutation is a change in DNA that can’t be repaired by the cell
Spontaneous mutations include replication errors (DNA polymerase insert incorrect nucleotide or mis-incorporated nucleotides persist after proofreading); and base modifications
Induced mutations are caused by chemicals and radiation which can result in point mutations, insertions, and/or deletions
What are the two kinds of point mutations (base substitutions)? What differentiates them?
Transition = purine to purine or pyrimidine to pyrimidine (A between G; T between C)
Transversion = purine to pyrimidine and vise versa (A to C or T; G to T or C; T to A or G; or C to A or G)
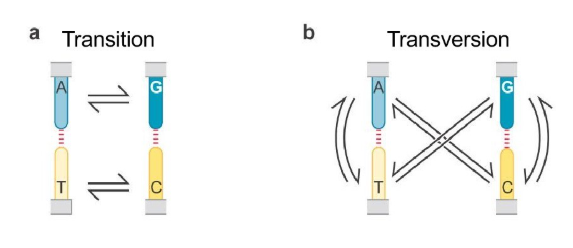
What causes point mutations?
Replication errors that weren’t fixed
Tautomeric forms of nucleotides
Damaged nucleotides (caused by alkylation, oxidation, radiation, and or depurination)
What are tautomers and what can they lead to?
Tautomers: alternate chemical forms that differ by only a single proton shift
tautomeric shifts may lead to permanent base-pair changes.
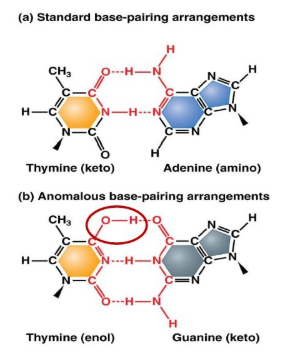
What is a fixed mutation?
A mutation that can’t be repaired
What is damage from alkylation, oxidation and deamination chemically look like?
oxidation = Chemically alters bases
Alkylation = Adds alkyl groups (CH3 or Ch3Ch3) to amino or keto groups in nucleotides
Deamination = Removes amino group, changes base; can be spontaneous or induced
Depurination = loss of nitrogenous base, usually a purine, any base may be inserted during replication
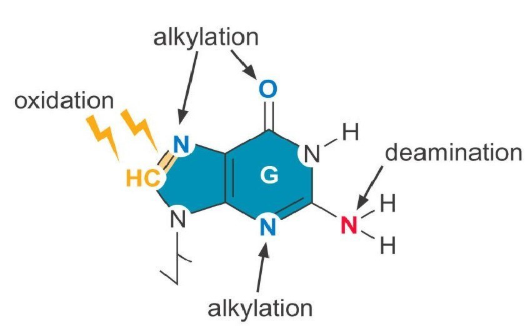
What are Mutagens-Base analogs?
Base analogs are chemicals that closely resemble normal DNA bases and can be mistakenly substitued for bases into DNA during replication
increase tautomeric shifts and sensitivity to UV light
What are missense, nonsense, and silent mutations?
Missense mutation: a substitution in the codon that codes for an amino acid is changed to a code for a different amino acid
Nonsense mutation: a substitution in a codon that codes for a amino acid is changed to a stop codon
Silent mutation: a substitution in a codon that codes for an amino acid that doesn’t change the amino acid

What are intercalating agents? What can it cause?
chemicals with dimensions and shapes that wedge between DNA base pairs which cause base-pair distortions and DNA unwinding which can lead to insertions and deletions (ex. ethidium bromide)
What are pyrimidine dimers and what are they caused by?
Pyrimidine dimers are a common product of UV light, where two thymine on the same strand of RNA will bind to themselves rather than adenine. the Dimers distort the DNA conformation and can inhibit normal replication
What does ionizing radiation cause? Which rays are considered mutagenic?
Mutagenic: X-rays, gamma rays, cosmic rays
penetrates deeply into tissues, causes ionization of molecules → free radicals
causes double strand breaks
Cells have multiple mechanisms to repair genetic damage. What ensures the high fidelity of DNA replication?
The fidelity of DNA replication comes from three mechanisms:
Accurate nucleotide selection by DNA polymerase
Immediate proofreading via 3′→5′ exonuclease activity
Post-replicative mismatch repair that corrects remaining errors
What are the four repair mechanisms that cells do to fix DNA problems before they create mutations?
mismatch repair
base excision repair
nucleotide excision repair
double-strand break repair
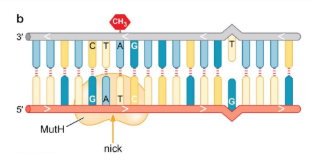
What are the DNA lesions fixed, causes of DNA lesions, what recognizes the DNA lesions, overall steps, and cell cycle stage most active in, for Mismatch repair?
Fixed = Mismatched single nucleotide variations (insertions n deletions)
Cause of DNA lesions = replication error that escaped DNA pol proofreading
recognized by = Distortions in the DNA helix caused by mismatched bases
general steps = 1. repair enzyme recognize mismatch, 2. MutH (endonuclease) makes a nick in the unmethylated DNA strand, 3. exonuclease unwinds and degrades the nicked DNA strand until the mismatch region is reached, 4. polymerase fills in gap, 5. ligase seals nick
Most active in = S phase
Notes = decreased with cancer and aging
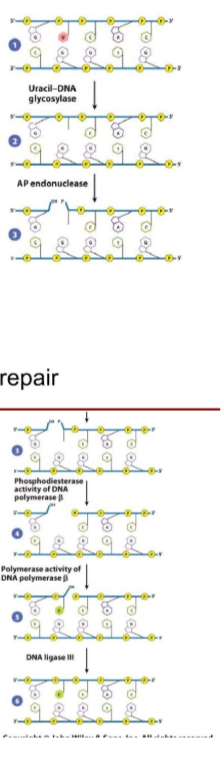
What are the DNA lesions fixed, causes of DNA lesions, what recognizes the DNA lesions, overall steps, and cell cycle stage most active in, for Base excision repair
Fixed = Altered or damaged bases that distort the DNA helix
Cause of DNA lesions = Deamination, oxidation, alkylation, methylation spontaneous base damage
recognized by = DNA glycosylases specific for particular altered bases
general steps = 1. DNA glycosylase removes damaged base; 2. Endonuclease cleaves DNA backbone; 3. DNA polymerase fills in correct nucleotide; 4. DNA ligase seals strand
Most active in = All throughout cell cycle
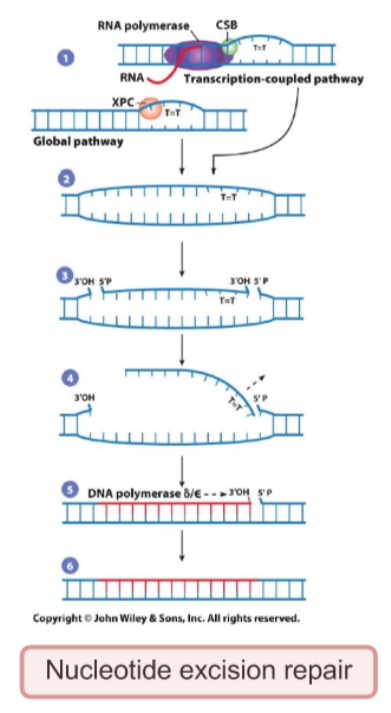
What are the DNA lesions fixed, causes of DNA lesions, what recognizes the DNA lesions, overall steps, and cell cycle stage most active in, for Nucleotide excision repair
Fixed =Bulky DNA lesions (e.g. pyrimidine dimers)
Cause of DNA lesions = UV radiation, chemical damage
recognized by = Transcription coupled-NER: stalled RNA pol with CSB protein; global genomic-NER: XPC protein
general steps: 1.Damage recognition by stalled DNA polymerase + CSB; 2. DNA unwinding by XPB and XPD (TFIHH) 3. Incision by XPG and XPF-ERCC1( on each side) 4.Excision of games ssDNA fragment 5. DNA polymerase δ/ε fills gaps and DNA ligase I seals strand
Most active in = entire cell cycle
Notes = Xeroderma pigmentosum is caused by mutations in NER-related genes
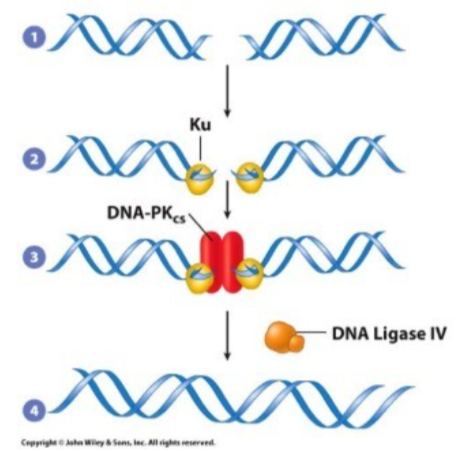
What are the DNA lesions fixed, causes of DNA lesions, what recognizes the DNA lesions, overall steps, and cell cycle stage most active in, for Non-homologous end joining (NHEJ)
Fixed = Double stranded breaks (no template)
Cause of DNA lesions = ionizing radiation and chemicals
recognized by = Lu proteins binding broken DNA ends
general steps = 1. Ku binds free DNA end 2. DNA dependent protein kinase catalytic subunit mediates repair 3. DNA ligase IV seals end
Most active in = throughout the cell cycle, primarily G1 and early G, when there are no sister chromatids
Notes = error prone, remember done WITHOUT template
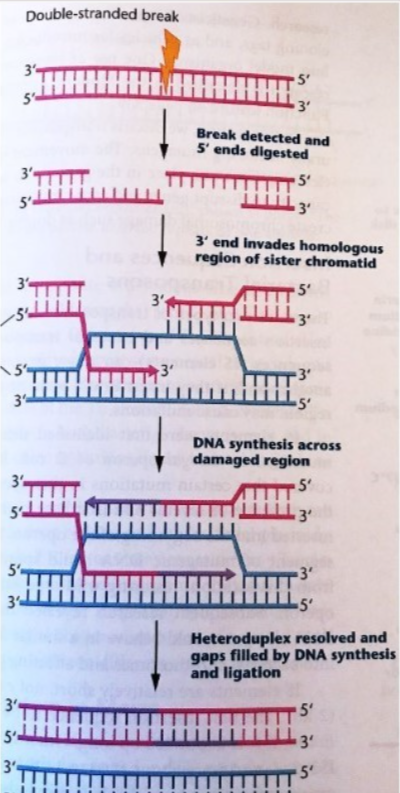
What are the DNA lesions fixed, causes of DNA lesions, what recognizes the DNA lesions, overall steps, and cell cycle stage most active in, for Homologous recombination?
Fixed = dsDNA breaks (template available)
Cause of DNA lesions = ionizing radiation, chemicals, replication fork collapse
recognized by = MRN complex? Using sister chromatid as template
general steps 1. Break detected, 2. Nuclease digests both DNA ends, 3. 3’end invades homologous region of sister chromatid, 4-5 polymerase synthesizes DNA across the damaged region, the DNA heteroduplex (holiday junction) is resolved, gaps filled in, nicks ligated
Most active in = Late S and G2
What happens simply in M phase and Interphase? About how long do these take?
M phase includes the process of mitosis and cytokinesis, lasts abt an hour or so
Interphase most of the cell cycle and may extend for days, week, or longer
What happens during G1 phase?
Cell growth and organelles replicate
What does the cell Check at G1/s checkpoint?
G1/S checkpoint = monitors size cell has achieved (big enough to divide?), evaluates condition of DNA (DNA integrity)
should i enter S phase or G0?
What happens during S (synthesis) phase?
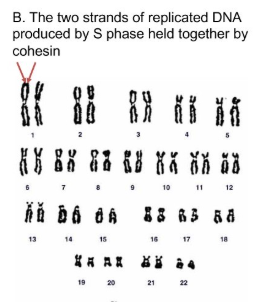
Each chromosome is duplicated for form 2 identical copies or sister chromatids of the chromosome
Each sister chromatid is held together by a protein complex called cohesion (green)
centromeres attach sister chromatids to each other and mitotic spindles attach there
In the cytoplasm, the centrosome (comprising a pair of centrioles) is duplicated too!
What are homologous chromosomes?
Each chromosome in the pair carriers the same set of genes with small sequence differences because one from mom one from dad
What are sister chromatids?
The two strands of replicated DNA produced by S phase held together by cohesin
What happens in G2 phase?
Continued growth in preparation for mitosis (gathering machinery)
What does the cell Check at G2/M checkpoint?
G2/M checkpoint = monitors if DNA replication is incomplete (pause cell if there is damage) monitors damaged DNA
Should I enter M phase or die?
What are the phases of Mitosis and about how long do they take?
Prophase = 36 mins
metaphase = 3 mins
Anaphase = 3 mins
Telophase = 18 mins
What happens during Prophase?
Mitotic spindle begins to form from the centrosomes
the DNA begins to condense
The nuclear envelope fragments then disappears
the single microtubules capture both of the sister chromatids on every homologous chromosome at the centromere
What happens in Metaphase plate? The M checkpoint is during this and what does it ask?
The captured chromosomes line up right between the two centrosomes
M checkpoint = monitors successful formation of spindle fibers system and attachment to kinetochores. Are both sister chromatids attached to opposite poles or centrioles?
What happens in Anaphase
the sister chromatids are ripped apart and pulled to opposite ends of the chromosomes to form the daughter chromosomes.
What happens in telophase vs cytokinesis?
telophase = nuclear division, mitotic spindle breaks down
cytokinesis = the dividing of the cytoplasm and splitting the cell
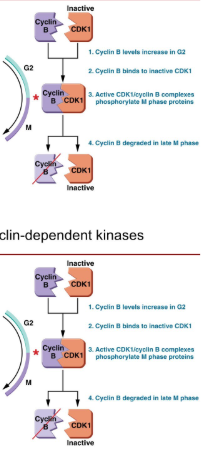
What regulates and mediates the progress of the cell cycle? How does it work?
Cyclins and Cyclin-dependent kinases (CDKs)
different cyclins are transcribed at different points in the cell cycle.
Phosphorylation and activation of proteins needed for the next phase.