Biochem Topic 1-4 (Exam 1)
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
23 Terms
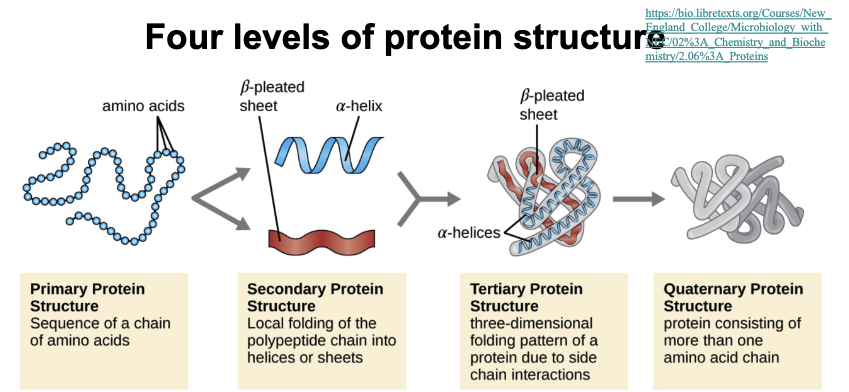
(Learning objective) Compare different levels of protein structure and how they relate to eachother
(learning objective) Describe what drives protein folding
Protein folding is entropy driven. Hydrophobic effect, water pushes non-polar (hydrophobic) parts of the protein away, causing them to hide inside the folded structure.
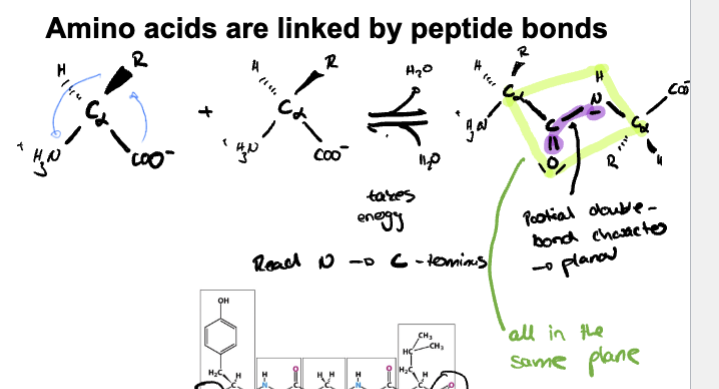
(learning objective) describe the fundamental properties of peptide bonds
-Covalent bond between carboxyl group and amino group on other amino acid
-double-bond character due to resonance on carbonyl group and C-N causing to be planar
-Polar
-Mostly exist in trans configurations
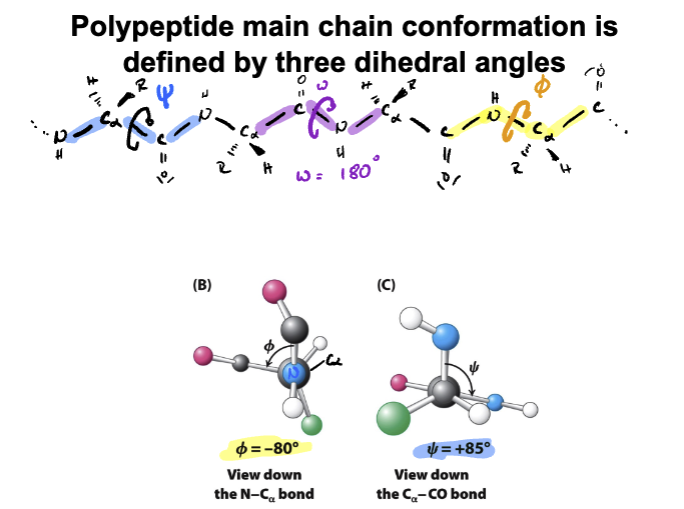
(learning objective) Describe the significance of the ramachandran plot
-Identifies possible steric combinations
-Identifies possible angles of rotation for backbone of protein
-Identifies secondary structure of an amino acid

1)Phi 2) Psi 3) omega (not pictured)
(learning objective) basic understanding of how proteins fold
1) Proteins start as long chain of amino acids (primary protein) Each amino acid has different properties some hydrophobic and some hydrophilic
2)hydrophobic effect kicks causing folding
3)back bone hydrogen bonding cause formation of alpha-helices or beta-sheets (local structure)
4) protein twists and folds into 3D shape which held by hydrogen bonds, ionic, disulfide, van der waals, and hydrophobic effect
(learning objective) Explain the conclusions of Anfinsen’s experiment
-Experiment that focused enzyme RNase to study tertiary structure and study ideal properties of formation
-Experiment 1- Urea and BME were added to enzyme where it was denatured. When Urea and BME were removed at the same time where enzyme re-formed into tertiary structure. (4-disulfide bonds)
-Experiment 2- Urea and BME were added to enzyme where it was denatured, but this time BME was removed first followed by Urea where enzyme was seen to be scrambled with improper di-sulfide bonds
-Experiment 3-Urea added with trace amounts of BME were added to scrambled enzyme where correct tertiary structure was re-formed
(BME helps break wrong di-sulfide bonds and helps form correct ones as protein folds)
(Urea helps disrupt non-covalent interactions that hold 3D structure together)
-Conclusion: Structure and function of protein are encoded in the primary sequence
Are amino acid known to be in Trans or Cis formation?
Amino acids are known to be in Trans formation with only glycine and proline being able to adopt Cis

How long would protein take if it tried every possible folding shape possible?
levinthal’s paradox- take longer than the age of the universe

Protein folding funnel model visualizes the folding process as an energy landscape. Top of the funnel unfolded protein has high entropy and energy, as folding progresses, entropy and energy decrease in a downward trend eventually reaching lowest point known as native state.
Global minima represents the overall lowest energy and entropy state known as the native state; while local minima (local valley) represents partially folded states that are kinetically trapped
Entropy overall represents the number of possible conformations a protein can adopt for example at the top entropy is high because unfolded proteins can exist in many shapes
Free energy drives folding process, starts from high free energy to native state with lowest free energy (most stable)
why are proteins marginally stable?
Good thing, conformation changes are important for function. Machines ultimately need to be able to move to do their jobs. Protein turn over is important.
Proteins need to be flexible to do their jobs, but stable enough to hold their shape
What is a “tag”
a short protein/sequence that we attach to the protein we want to study
Define isoelectric point
PH at which net change of protein is 0
(learning objective) Basic understanding of how proteins fold.
proteins fold step-by-step into a specific shape using weak bonds and interactions.
Primary structure: chain of amino acids made during translation (in ribosome)
Secondary structure: Parts of the chain form alpha helices and beta sheets held by hydrogen bonds
Tertiary structure: The whole chain folds into a 3-D shape stabilized by hydrophobic interactions, hydrogen bonds, ionic bonds, and disulfide bridges.
Quaternary structure: Some proteins have multiple folded chains that come together to form a final protein.
(learning objective) Describe the fundamental properties of peptide bonds
Bond between a Carboxylic acid group of one amino acid and NH2 of another amino acid forming a peptide bond that has partial double bond characteristics because of resonance.
planar and rigid so it doesn’t rotate freely
Trans configuration favored
(learning objective) Describe the significance of the ramachandran plot
Organizes possible steric combinations
Tells us about secondary structures, helps identify alpha-helices and beta sheets in different regions
Shows allowed angles of rotation for the backbone of a protein
Phi=Nitrogen to Alpha carbon
Psi=Alpha carbon to carbon bond
omega bond= rotation around peptide bond (usually fixed at 180 degrees (trans))
(learning objective) Describe what drives protein folding
Entropy driven by hydrophobic effect, non-polar side chains avoid water becoming buried inside the folded protein structure.
(learning objective) Explain the conclusions of Anfinsen’s experiment
Describe Anfinsen’s experiment
-Ran experiments using denaturing agents Urea (disrupts non-covalent bonds) and BME (breaks disulfide bonds) that focused on an enzyme called RNase to study tertiary structure and study proper conditions it was formed
-Experiment #1- Excess Urea and BME added together where it denatured enzyme. When re-added enzyme tertiary structure was re-formed
-Experiment #2- Excess Urea and BME added together where first B. Merca was removed and then Urea where then a scrambled enzyme was seen with improper disulfide bonds
Experiment #3-Trace amounts of BME. after removing Urea, were added to the scrambled enzyme where the correct tertiary structure was re-formed.
Three experiments combined showcased that the structure and function of a protein are encoded in the primary sequence.
(mnemonic) For hydrophobic
GAVLIMP
Glycine
Alanine
Valine
Leucine
Isoleucine
Methionine
Proline
(mnemonic) For aromatic
WYF
Tryptophan
Tyrosine
Phenylalanine
(mnemonic) For hydrophilic
STQNC
Serine
Threonine
Glutamine
Asparagine
Cysteine
(mnemonic) For negative charge/acidic
Ed
Glutamic acid
Aspartatic acid
(mnemonic) For Positive charge/basic
HKR
Histidine
Lysine
Arginine