11 Regulation of Gene expression
1/70
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
71 Terms
What are the three main types of epigenetic mechanisms?
Epigenetic mechanisms include CpG (DNA) methylation, Histone modification, and Regulatory ncRNAs.
What do epigenetic mechanisms establish regarding gene expression?
Epigenetic mechanisms establish potentially heritable changes in gene expression without altering the underlying nucleotide sequence.
How do epigenetic mechanisms influence gene expression without changing the DNA sequence?
They involve chemical alterations to chromatin that either enhance or reduce gene expression, without changing the DNA sequence.
What are histones described as?
Histones are highly conserved, alkaline, positively charged proteins.
Who first discovered histones and when?
Histones were first discovered by Albrecht Kossel in 1884.
What is a nucleosome?
A nucleosome is a combination of histone proteins (octomeres) that wrap DNA around them.
Who pioneered research correlating histone modifications with gene expression control, and what modifications did he identify?
Pioneering research by Vincent Allfrey (early 1960s) found that histones are modified by post-translational acetylation and methylation, correlating these modifications with gene expression control.
What is the effect of histone modifications on chromatin?
Histone modifications influence chromatin accessibility.
What state of chromatin is associated with methylation and gene inactivity?
Heterochromatin is associated with significant amounts of methylation, and in this state, the target gene is not expressed, meaning there is no accessibility.
What state of chromatin is associated with acetylation and gene activity?
Euchromatin is associated with acetylation on histone tails; higher acetylation leads to more relaxed chromatin, opening up critical regions for transcription factors to bind.
What are examples of critical regions in the genome that open up due to euchromatin formation?
Examples include enhancer elements and promoter regions.
What is DNA methylation mechanism?
DNA methylation refers to the addition of a methyl group to the DNA cytosine residues at the fifth carbon position (5mC; 5-methylcytosine).
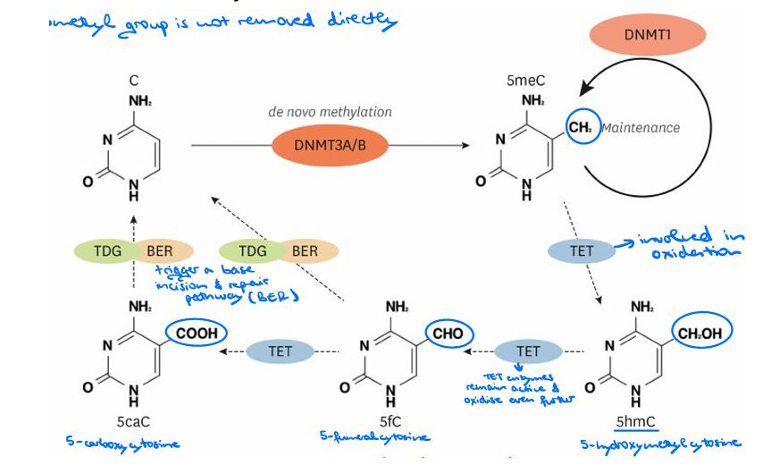
Where is DNA methylation commonly found in eukaryotes?
It is a common epigenetic mark in many eukaryotes, often found in the sequence context CpG (cytosine followed by guanine in 5’ to 3’ direction).
What are DNMT3A and DNMT3B, and what is their function?
DNMT3A and DNMT3B are DNA Methyltransferases (DNMTs) that mediate de novo DNA methylation, establishing initial patterns.
Where are DNMT3A and DNMT3B particularly abundant?
DNMT3A is abundant in immune cells, while DNMT3B is abundant in stem cells.
What is DNMT1, and what is its role?
DNMT1 is a DNA Methyltransferase that sustains the established methylation pattern, acting as a maintenance methyltransferase integral for the life of a multicellular organism.
What molecule donates the methyl group during DNA methylation?
S-adenosyl-L-methionine (SAM) donates the methyl group.
How does demethylation occur, and what enzymes are involved?
Demethylation involves TET (ten-eleven translocation) methylcytosine dioxygenases, which do not remove the methyl group directly but rather involve oxidation, as the enzymes remain active and oxidize even further.
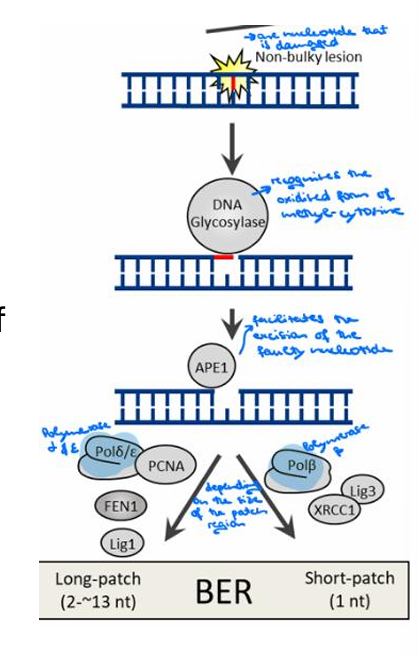
What type of DNA damage does Base Excision Repair (BER) correct?
BER corrects DNA damage from oxidation, deamination, and alkylation.
How is BER initiated?
BER is initiated by one of at least 11 distinct DNA glycosylases that recognize and remove the damaged base, depending on the type of lesion.
What is left after the damaged base is removed in BER, and how is it further processed?
An abasic site is left, which is further processed by short-patch or long-patch repair that largely uses different proteins to complete BER.
What does BER protect against?
BER protects against cancer, aging, and neurodegeneration.
Where does the BER process take place?
The process takes place in both nuclei and mitochondria.
What are the three main steps in the BER process?
The steps are Recognition (by DNA glycosylate), Excision (facilitated by APE1), and Resynthesis (by Polymeraseβ and Polymeraseδ/ξ).
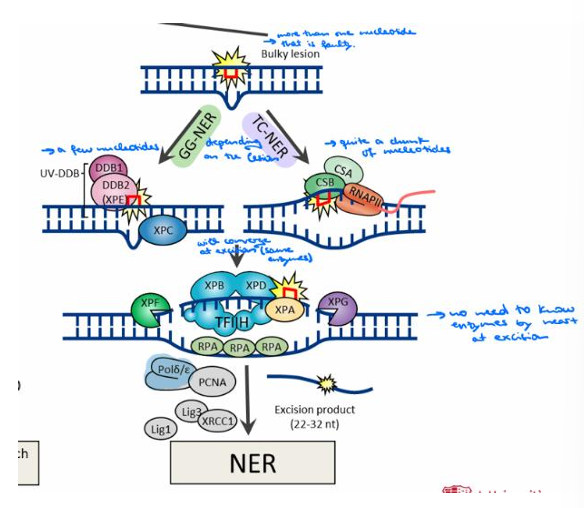
What is Nucleotide Excision Repair (NER) famous for in humans?
NER is famous for its unique ability in humans to remove photolesions produced by UV radiation (sun exposure).
How do NER factors detect damaged DNA?
NER circumvents direct lesion recognition; instead, NER factors detect unpaired single-stranded DNA opposite the damaged strand.
What are the two sub-pathways of NER, and what do they target?
The two sub-pathways are Global Genome NER (GG-NER), which eliminates lesions throughout the whole genome (a few nucleotides), and Transcription-Coupled NER (TC-NER), which eliminates lesions in the transcribing strand of active genes (quite a chunk of nucleotides).
What are the consequences of hereditary mutations in NER-associated genes?
Hereditary mutations in NER-associated genes are nonlethal but associated with UV sensitivity and cancer predisposition.
What are the three main steps in the NER process?
The steps are Recognition (using GG-NER or TC-NER depending on the lesion), Excision (where both processes converge using the same enzymes), and Resynthesis.
What type of DNA lesions does DNA Mismatch Repair (MMR) primarily fix?
MMR mainly repairs DNA lesions caused by faulty DNA replication or repair, addressing mismatches, small insertion and deletion loops, or deamination of 5-methylcytosine.
What is the critical role of MMR?
MMR plays a critical role in ensuring the fidelity of DNA replication.
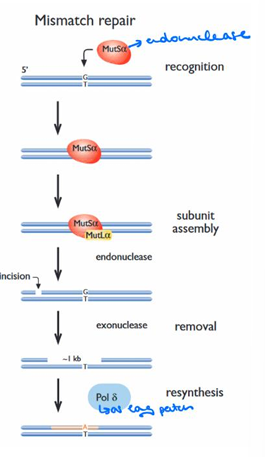
How is a mismatch recognized in the MMR process?
A mismatch is recognized by the MSH2•MSH6 heterodimer (MutSα) along with MLH1•PMS1 (MutLα).
What initiates the repair in MMR after mismatch recognition?
Repair is initiated by activating exonuclease-mediated degradation of DNA from a nick that is distant to the mismatch.
How is the degradation directed in MMR, and by what mechanism?
A hemi-methylated dGATC site (5′ or 3′ to the mismatch) is located and cleaved by the concerted action of MutS, MutL, MutH, and ATP, and degradation continues until the mismatched base is removed.
What fills in the resulting long excision tract in MMR?
The resulting long excision tract is filled in by DNA polymerase δ, inserting the correct nucleotide.
What is mutagenesis?
Mutagenesis is the formation of mutations in DNA molecules, which can be changes in DNA sequence or rearrangement of chromosomes.
What are spontaneous mutations, and why are they important?
Spontaneous mutations occur as 'mistakes' during DNA replication or mitosis and are essential for producing genetic variation necessary for natural selection.
What causes induced mutations?
Induced mutations result from environmental exposure to genotoxins (chemicals that alter DNA structure).
What are the concerns regarding mutations?
Mutations may lead to irreversible effects that can affect the fitness of organisms and, in turn, affect population-level processes.
What are Double-Strand Breaks (DSBs) considered in relation to chromosomal aberrations?
DSBs are considered critical primary lesions in the formation of chromosomal aberrations.
How can DSBs be induced or occur?
DSBs can be induced by exogenous agents (e.g., ionizing radiation) or occur spontaneously during cellular processes at significant frequencies.
What are unresolved DSBs implicated in?
Unresolved DSBs are implicated in various human disorders and cancers.
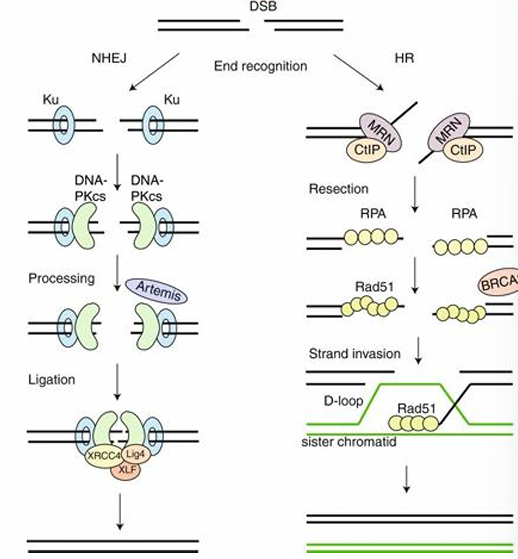
What are the two main repair pathways for DSBs, and what is their error status?
The two main repair pathways for DSBs are Non-Homologous End Joining (NHEJ), which is error-prone, and Homologous Recombination (HR), which is error-free.
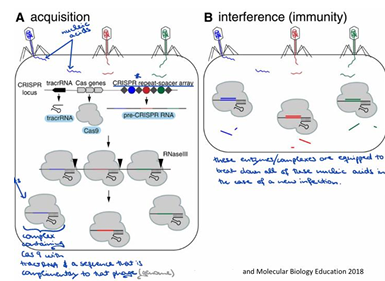
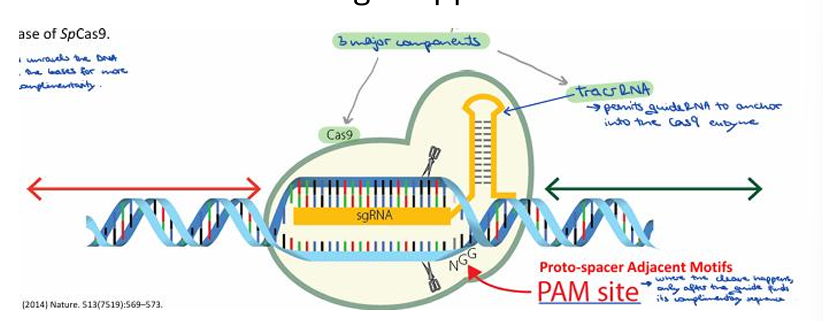
When was the CRISPR-Cas9 system discovered for genome modification, and by whom?
In 2012, George Church, Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang discovered that the CRISPR-Cas9 system can be used as a "cut-and-paste" tool to modify genomes by designing guide RNA to target specific regions.
What is the original nature and function of the CRISPR-Cas9 system?
It is a bacterial and archaeal immune system adapted for eukaryotic gene editing, used by prokaryotes as a defense mechanism against viral and plasmid cellular invaders.
How do prokaryotes use CRISPR-Cas9 to generate immunity?
They integrate DNA sequences identical to past invaders into their genome to generate immunity, for example, protection against blue, red, or green phages.
What are the main components of the CRISPR-Cas system?
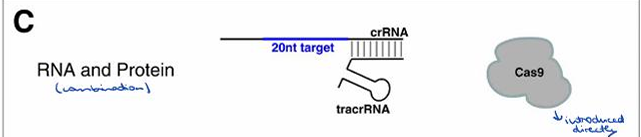
The components include CRISPR, Cas proteins (CRISPR-associated Proteins), Cas9, TracrRNA, and Pre-CRISPR RNA.
What is Cas9, and what is its function?
Cas9 is an RNA-guided endonuclease that cleaves DNA at sequences that bind to the crRNA of the Cas9 RNP.
Describe the "Acquisition" process in CRISPR-Cas system immunity.
Phages inject their nucleic acid into the bacterium; these then form a complex containing Cas9, with tracrRNA and a sequence complementary to that phage (genome).
Describe the "Interference (immunity)" process in CRISPR-Cas system.
These enzymes or complexes are equipped to break down all of these nucleic acids in the case of a new infection.
How many types of CRISPR-Cas systems have been identified in bacteria and archaea?
Three different types have been identified: Type I, Type II, and Type III.
What is the first step in generating a desired mutation using CRISPR-Cas9?
The first step is the design of the guide RNA.
What must the 20-nucleotide target region of the guide RNA be adjacent to for SpCas9?
For SpCas9, the 20-nucleotide target region of the guide RNA must be adjacent to a PAM site, specifically 5’-NGG-3’.
What is the function of the PAM (Proto-spacer Adjacent Motifs) site in CRISPR-Cas9?
The PAM site indicates where the cleavage happens.
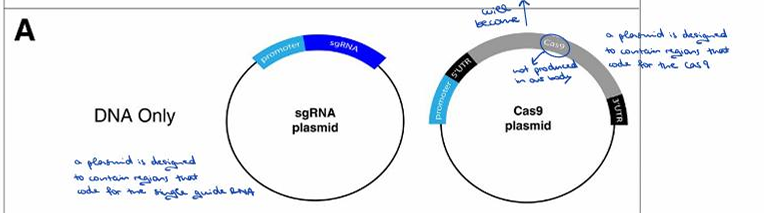
What is a concern when introducing foreign nucleic acid into a cell for CRISPR/Cas9 components?
If a foreign nucleic acid is introduced into a cell, the innate immune system attacks it.
Describe the "DNA only" method for introducing CRISPR/Cas9 components.
One plasmid is designed to code for the single guide RNA, and another plasmid is designed to code for the Cas9 protein (as it is not produced in our body).
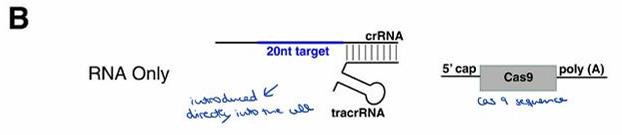
Describe the "RNA only" method for introducing CRISPR/Cas9 components.
TracrRNA is introduced directly into the cell, and the internal machinery of our cells will produce these molecules.
Describe the "RNA and protein" method for introducing CRISPR/Cas9 components.
Both tracrRNA and Cas9 protein are introduced directly into the cell.
What are "off-target effects" in CRISPR-Cas9 application?
Off-target effects are the introduction of unintended mutations or genetic changes, where CRISPR will cut and cleave wherever it feels like it, and studies show these can far outweigh intended ones.
What is the concern about "heritability" with CRISPR-Cas9?
If gene editing occurs in germline cells (sperm or eggs), changes could be passed to future generations, potentially forming unintended mutations or other genetic changes.
What are "unintended physiological consequences" of CRISPR-Cas9 application?
There is a risk of unforeseen health problems such as increased cancer risk or autoimmune disorders due to effects on the immune system or other physiology.
What general approaches are recommended for mitigating potential unintended long-term effects of CRISPR-Cas9?
Careful monitoring and regulation, rigorous testing, and close monitoring of long-term effects are needed.
Name two specific strategies for monitoring gene expression to mitigate CRISPR-Cas9 effects.
Techniques like quantitative PCR or RNA sequencing can be used to track changes over time.
Name two specific strategies for monitoring genome integrity to mitigate CRISPR-Cas9 effects.
Techniques such as whole-genome sequencing or karyotyping for larger regions can be used to ensure edited cells do not develop unintended mutations.
How can high-precision CRISPR-Cas9 systems help mitigate unintended effects?
They are designed for greater specificity and accuracy to reduce unwanted changes.
How do Anti-CRISPR-Cas9 proteins help mitigate off-targeting?
They inactivate Cas9 protein after target site cleavage to reduce off-targeting (so it doesn’t cleave any other parts of the genome).
How can bioinformatics tools help mitigate CRISPR-Cas9 concerns?
Better methods can be developed to design more accurate guide RNA and predict off-targeting.
Is there a single "gold standard" method for mitigating CRISPR-Cas9 long-term effects?
No, a combination of these strategies is recommended.
Process of MMR
What does CRISPR stand for?
Clustered regulatory interpaced short palindromic repeats
CRISPR mechanism
The CRISPR mechanism involves a system used by bacteria to defend against viral infections, utilizing RNA to guide the Cas9 protein in cutting specific DNA sequences.