Lecture 30: Protein translocation and folding in the ER
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
what is the name of the ER translocator
Sec61
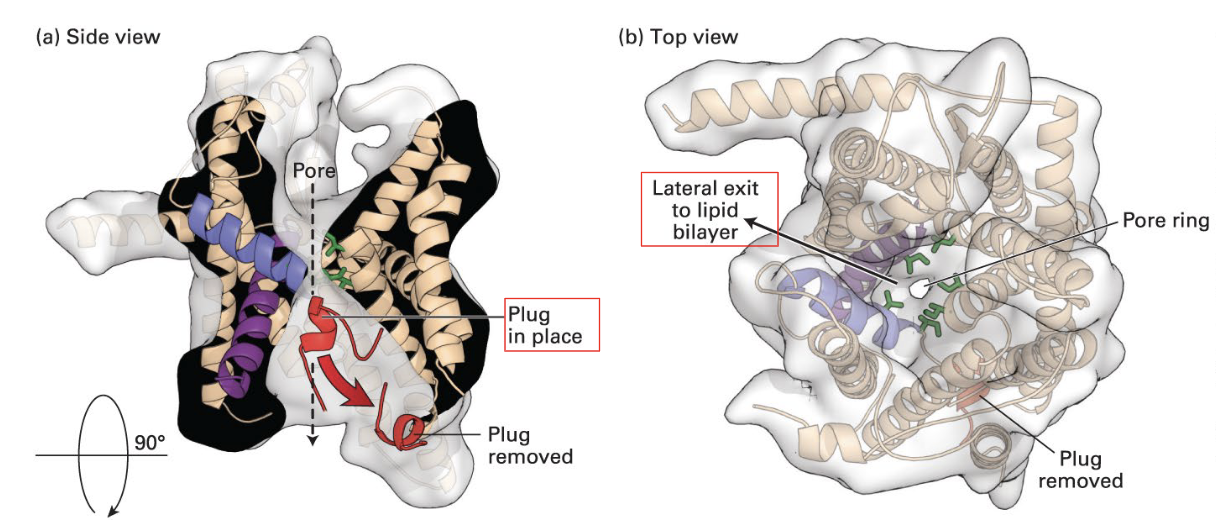
structure of Sec61
pore ring (isoleucines) in the middle of the structure where the peptide can pass through, and it prevents leakage of ions and other small molecules when the polypeptide is translocated. There is a small peptide that acts as a plug when the polypeptide is absent.
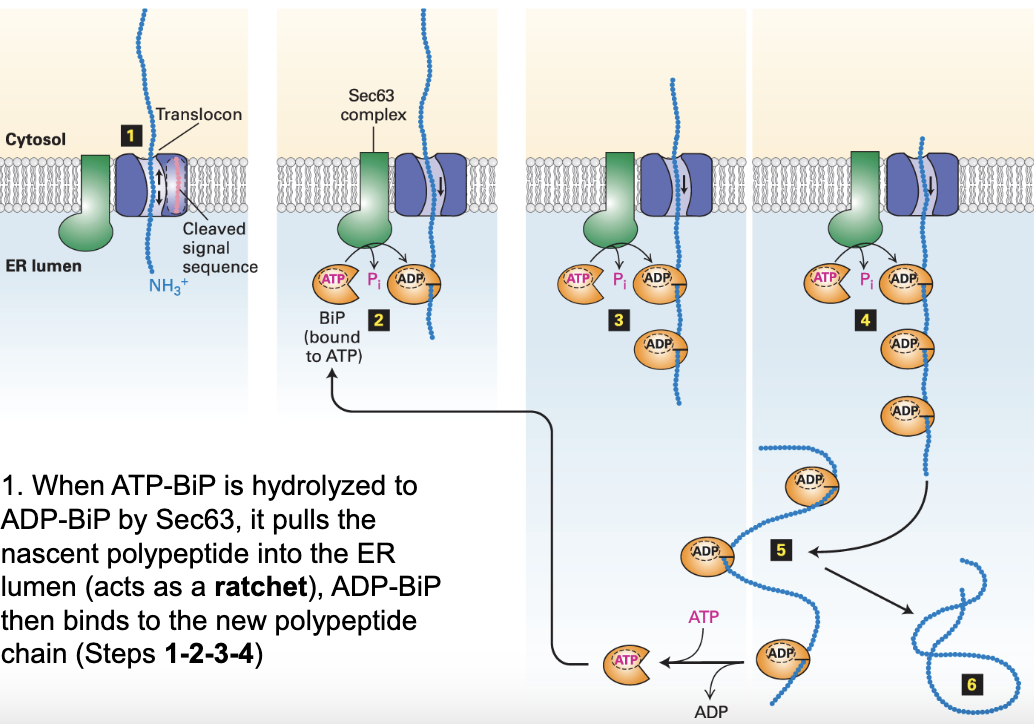
what is BiP?
Binding of immunoglobulin Protein is an ATPase that exists in the lumen of the ER and is used to help protein folding
ADP-BiP in protein folding
ATP-BiP is hydrolyzed to ADP-BiP by Sec63, and pulls the nascent polypeptide into the ER lumen. Many ADP-BiPs bind
Bound ADP-BiPs prevent non-specific aggregation of nascent polypeptides (so they can be folded correctly)
Once the proteins are folded correctly ADP-BiPs are phosphorylated back to ATP-BiP and are released
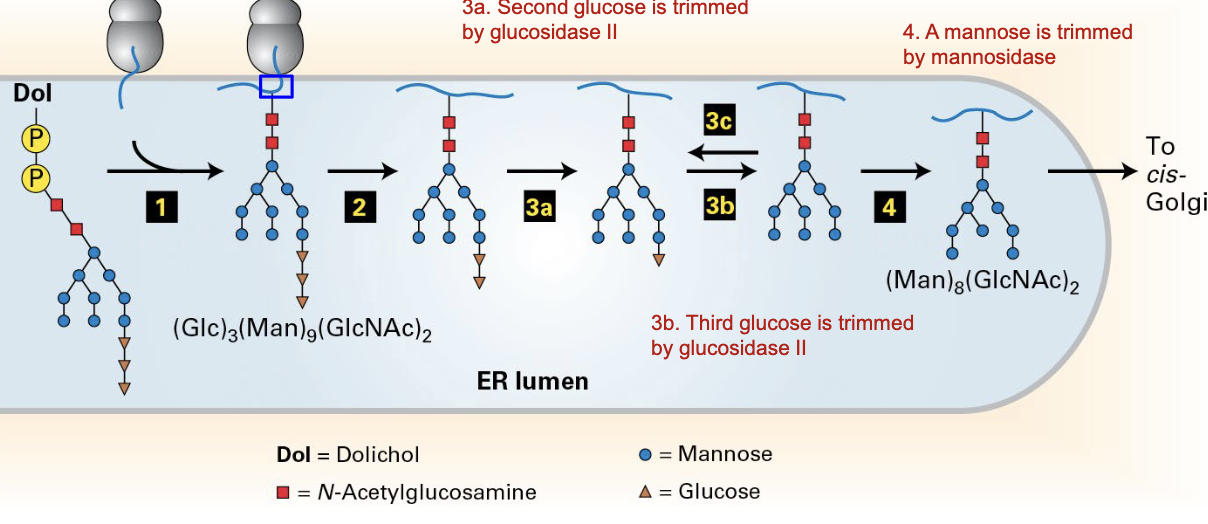
N-glycosylation of proteins in ER
pre-formed oligosaccharide (with three glucoses and 9 mannoses) is added to N-terminus of nascent protein
first glucose cleaved by glucosidase I
second glucose cleaved by glucosidase II
a) third glucose cleaved by glucosidase II if folded properly
b) new glucose added back by glucosyl-transferase if not folded properly
mannose is cleaved by mannosidase, and the remaining complex is sent to the Golgi
what happens once the first two glucoses are removed from the N-glycosylation complex?
the third glucose is recognized by chaperone proteins (CNX/CRT) for folding: incorrectly folded ones are trapped and off-pathway is prevented
what happens when the 3rd glucose is removed from the N-glycosylation complex?
removal of the third glucose releases the protein form the chaperone proteins (CNX/CRT)
what happens if the protein cannot fold properly after a long time?
more mannoses are trimmed and the complex is recognized for degradation by OS-9
what do disulfide bonds by PDI do?
stabilize folded protein structure
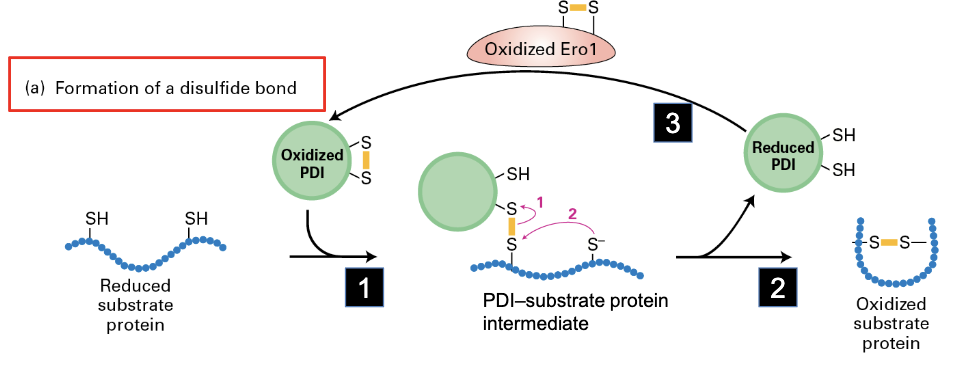
mechanism for formation of disulfide bond
reduced substate protein interacts with oxidized PDI to form a disulfide bond with itself, and reducing PDI in the process. this PDI gets converted back to oxidized form via Ero1
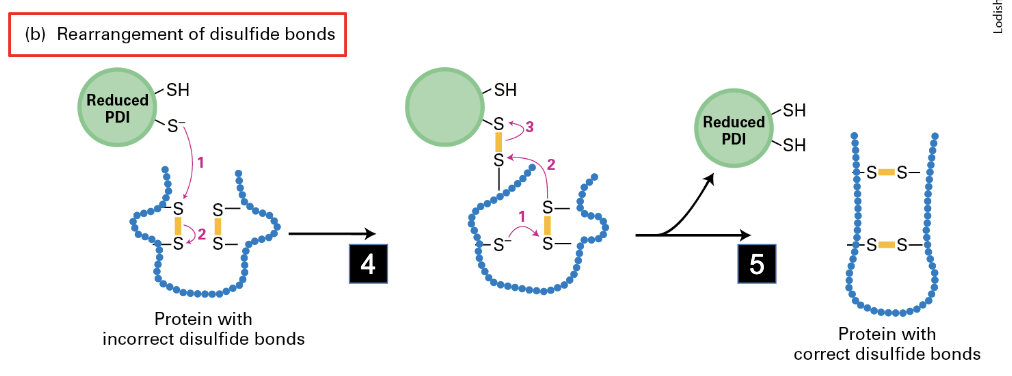
mechanism for rearrangement of disulfide bonds
reduced PDI interacts with incorrect disulfide bonds to rearrange electrons allowing for correct bonds
what is a type IV membrane protein?
a membrane protein that has more than one transmembrane domain (TMD)
what structure do TMDs have?
alpha-helices, with 18-22 amino acids (with the majority of them being hydrophobic)
example of type IV TMDs
Sec61, GPCRs
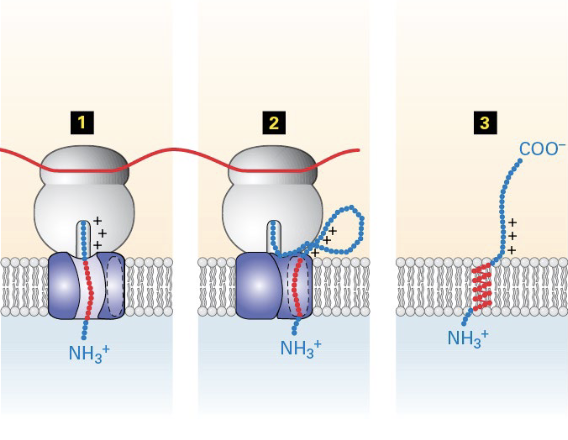
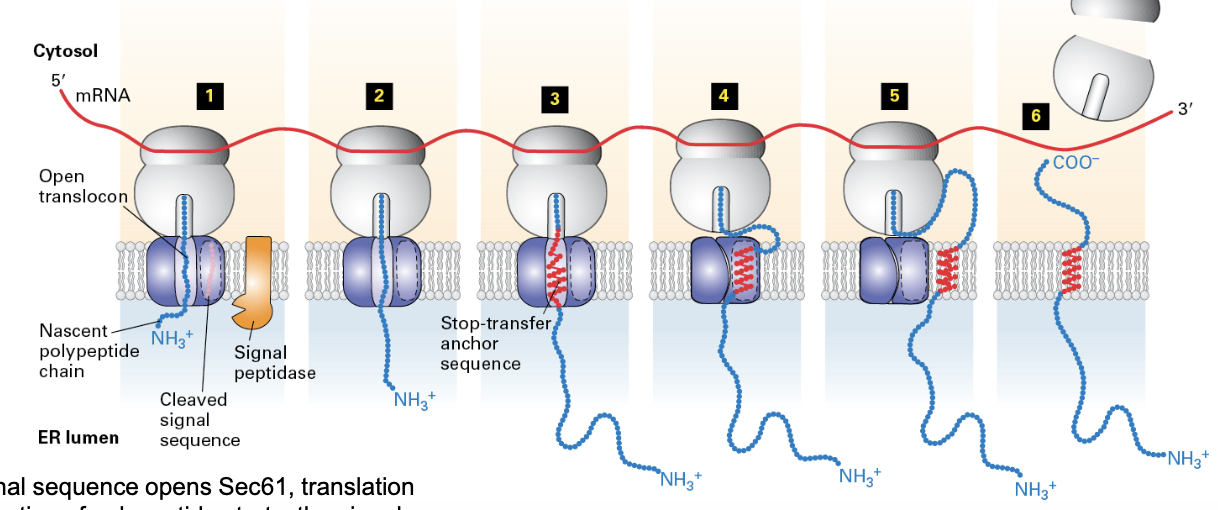
insertion of type I membrane proteins to ER membrane
ER signal sequence opens Sec61, translation and translocation of polypeptide begins, signal is cleaved and degraded
translocation stops when the TMD is met
translation continues in the cytosol, TMD is moved laterally to Sec61 (to lipid bilayer)
ribosome released when translation is completed, leaving protein anchored to the membrane
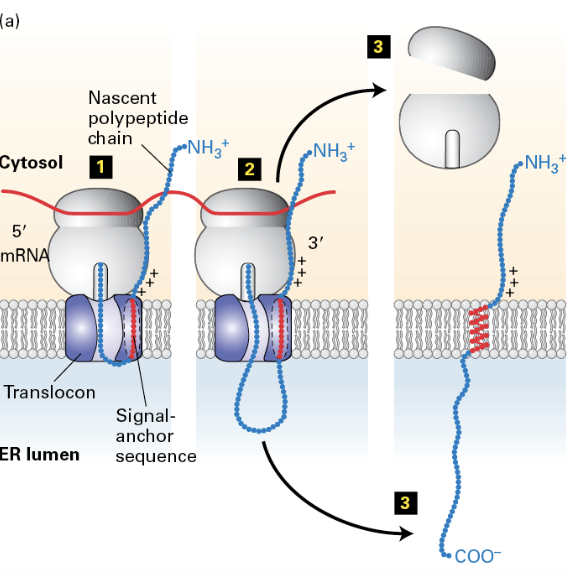
how does Sec61 open to allow the TMD to exit?
like a clam shell
insertion of type II (N-cytosolic, C-lumenal) membrane proteins
no N-terminus signal sequence so the TMD acts as an internal anchor sequence that directs the complex to the ER and opens the translocator (N-terminus stays in cytosol)
translation and translocation of polypeptide behind TMD continues
ribosome released when translation is done, insertion of C-terminus into the ER lumen
insertion of type III (N-luminal, C-cytosolic) membrane proteins
TMD enters the translocator in orientation such that the C-terminus remains in the cytosol and the N-terminus is in the lumen
translation occurs in cytosol
ribosome release when translation is done