BIOL 2030: Module 10
1/92
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
93 Terms
Chromosome Variations
permanent chromosomal changes
can be passed on to offspring if they occur in cells that will become gametes
germline cells
two general types of chromosomal variation:
chromsome rearrangement
variation in chromosome numbers
Chromosome Rearrangement
changes in the structure of individual chromosomes
4 types:
deletions
duplications
inversions
translocation
Variation in Chromosome #
changes in the # of chromosomes
1 or more individual chromosomes are added or deleted
Deletions
type of chromosomal rearrangement in which there is a loss of a segment, either internal or terminal, form a chromosome
arise by:
terminal ends breaking off (one break)
internal breaking and rejoining of incorrect ends (two breaks)
unequal crossing over
major effect: loss of genetic information
importance depends on what, and how much is lost

Detection of Deletions
deletion loops can be detected during meiosis
also by a variety of molecular methods that detect lower heterozygosity or gene dosage
Consequences of Deletion
loss of DNA sequences
deletions that span a centromere result in an acentric chromosome that will most likely be lost during cell division, which may be lethal
deletion can allow for the expression of alleles that are normally recessive
pseudodominance
deletions can affect gene dosage
when a gene is expressed, the functional protein is normally produced at the correct level or dosage
some (not all) genes require two copies for normal protein production
if one copy is deleted, a mutant phenotype can result
phenotypic effects depend on the size and location of deleted sequences
cri du chat
Acentric Chromosome
chromosome which lacks a centromere
Psuedodominance
genetic phenomenon where a recessive trait appears to be dominant, often because the masking dominant gene has been deleted
deletions can allow expression of alleles that are normally recessive
Haploinsufficiency
when one working copy (allele) of a gene isn't enough to produce the normal amount of protein needed for a healthy function, leading to a genetic disorder, even though the other gene copy is fine
some, though not all, genes require two copies of normal protein production.
if one copy is deleted, a mutant phenotype can result
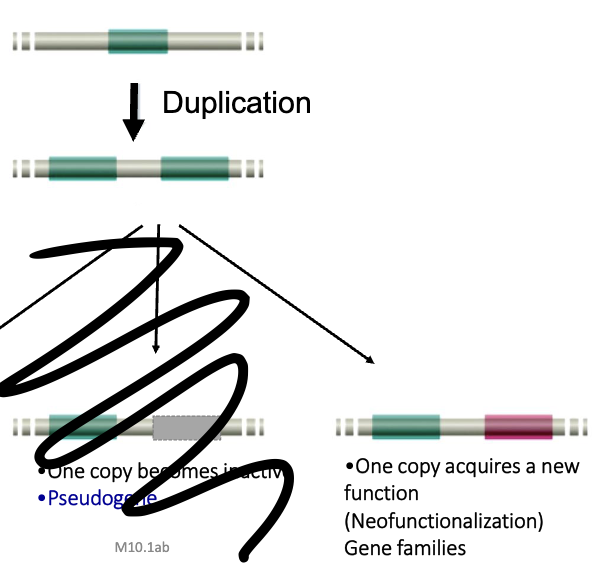
Duplications
repetition of a chromosome segment
simplest form: tandem duplication
a single gene, or cluster of genes can be duplicated. nothing has been lost, so duplications often have little or no effect on phenotype/viability
offspring with duplications are usually viable
however, in some cases, excess or unbalanced dosage of gene products resulting from duplications can cause problems
very important in evolution, because extra copies of genes provide raw material for new genes and adaptations
about 5% of all human genome consists of duplications
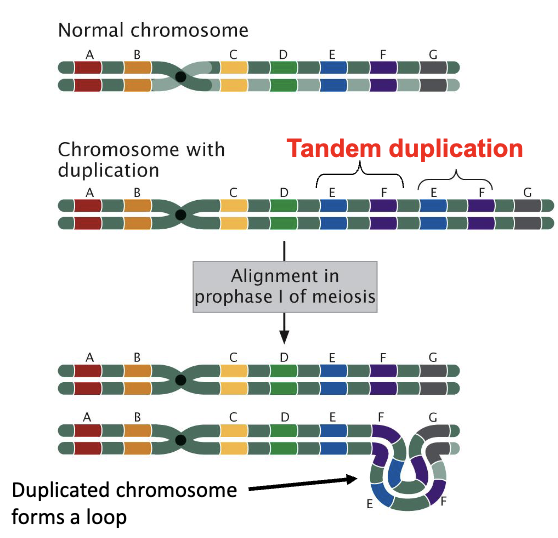
Tandem Duplications
a type of chromosomal mutation where a DNA segment is copied and inserted immediately next to the original sequence, creating a "head-to-tail" arrangement
ex. ABC becomes ABCBC
Origin of Duplications
unequal crossing over of misaligned chromosomes during meiosis generates duplications, and deletions
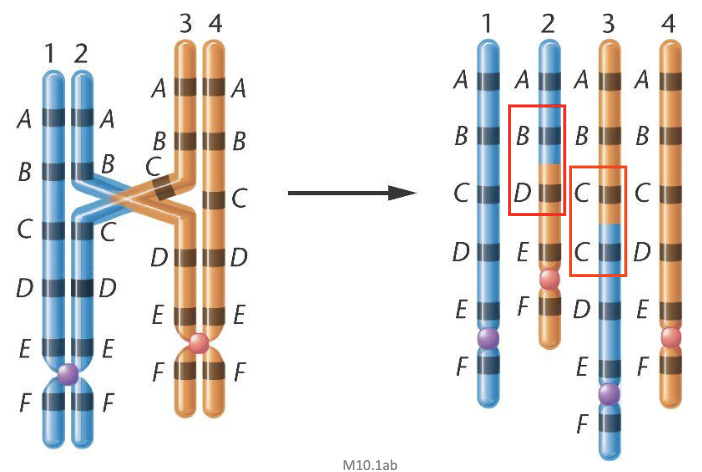
Detection of Duplications
duplicated chromosomes (specifically in the case of tandem duplication) forms a loop during prophase I of meiosis
also by various molecular methods that detect higher gene dosage
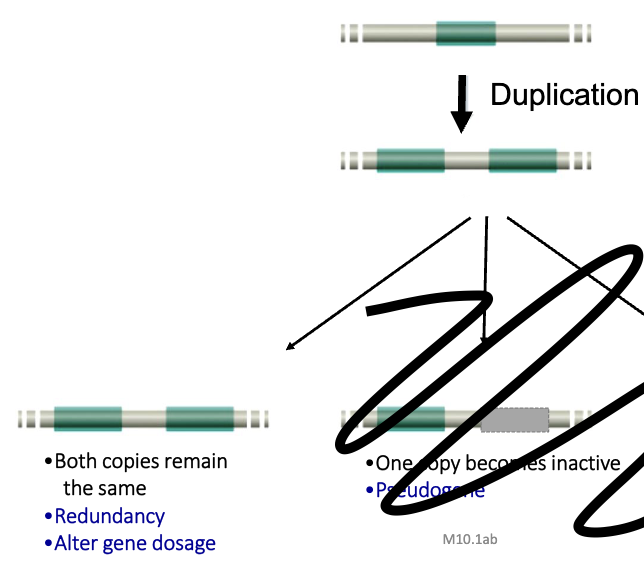
Consequences of Duplication
3 possibilites:
redundancy
pseudogene
neofunctionalization
gene dosage may affect phenotype
amount of protein synthesized is often proportional to the # of gene copies present, so extra genes can lead to excess proteins
ex. bar region in Drosophila represent that the more copies of a gene (X chromosome), the fewer eye facets
Redundancy
both copies remain the same post duplication
alters gene dosage
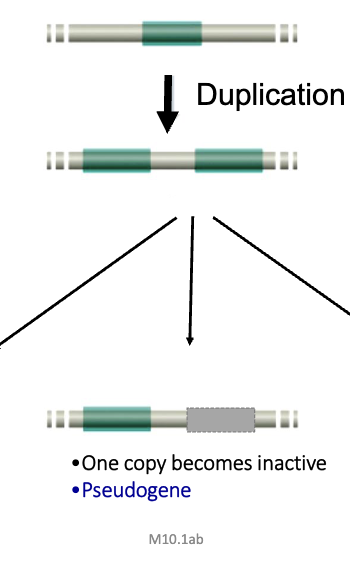
Pseudogene
one copy becomes inactive after duplication
Neofunctionalization
after duplication, one copy acquires a new function
source of new genes, and multigene families
ex. globin gene family
Inversions
two breaks on a chromosome followed by reinsertion in the opposite orientation causes inversion
pericentric inversions: around
paracentric inversion: beside or beyond
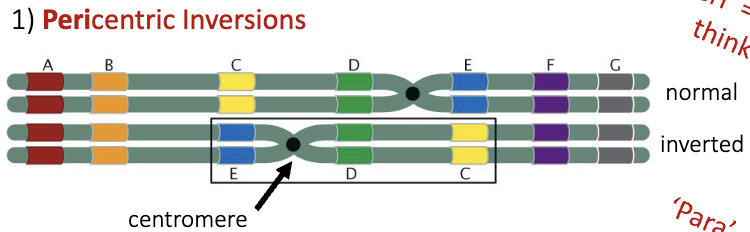
Pericentric Inversions
“peri” = around
think perimeter
chromosomal rearrangement where a segment of a chromosome that includes the centromere is broken, flipped, and reinserted in reverse order. This changes the order of genes on the chromosome, but not the total amount of genetic information
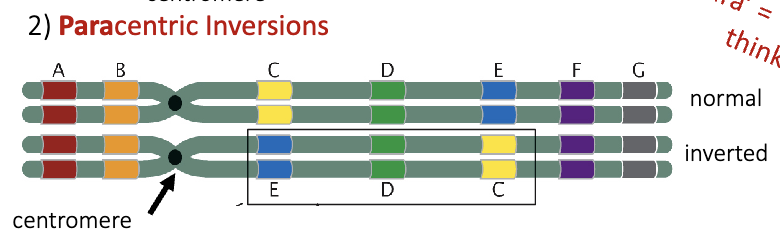
Paracentric Inversion
“para” = beside or beyond
think paranormal
a type of chromosomal rearrangement where a segment of a chromosome breaks, flips, and reinserts itself without including the centromere. This results in both breaks occurring on the same arm of the chromosome
Inversion Effect on Phenotype
often, no effect
however, sometimes there is an effect driven by the change in the position of the genes
Inversion Consequences
position effects:
change in position can sometimes alter expression
genes in/near chromatin may not be expressed
suppression of recombination:
if no crossing over occurs, gametes produced are usually viable because genetic information is not lost or gained
if crossing over occurs OUTSIDE of the inverted region, the gametes are viable.
if crossing over occurs INSIDE of the inverted region, some nonviable gametes and reduced recombination frequency
Crossing Over Within a Paracentric Inversion
crossing over between inverted and non-inverted chromosomes
C and D
formation of an inversion loop, and a crossing over occurs within the inversions
see crossing over between inverted and non-inverted chromosome between C and D
formation of a dicentric bridge
resulting in:
reduced recombination frequency
reduced fertility
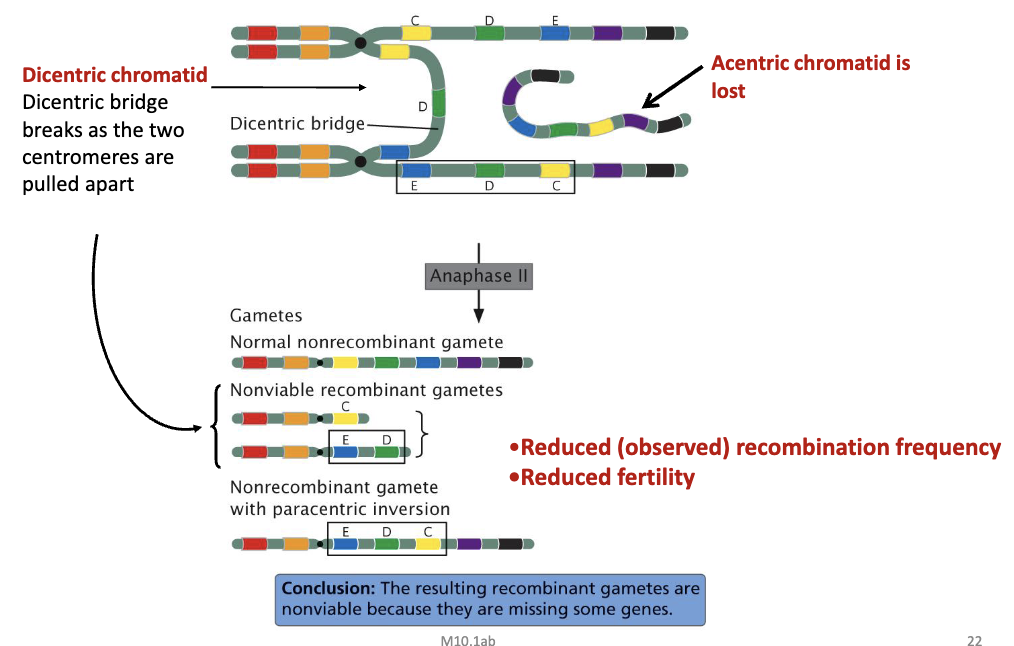
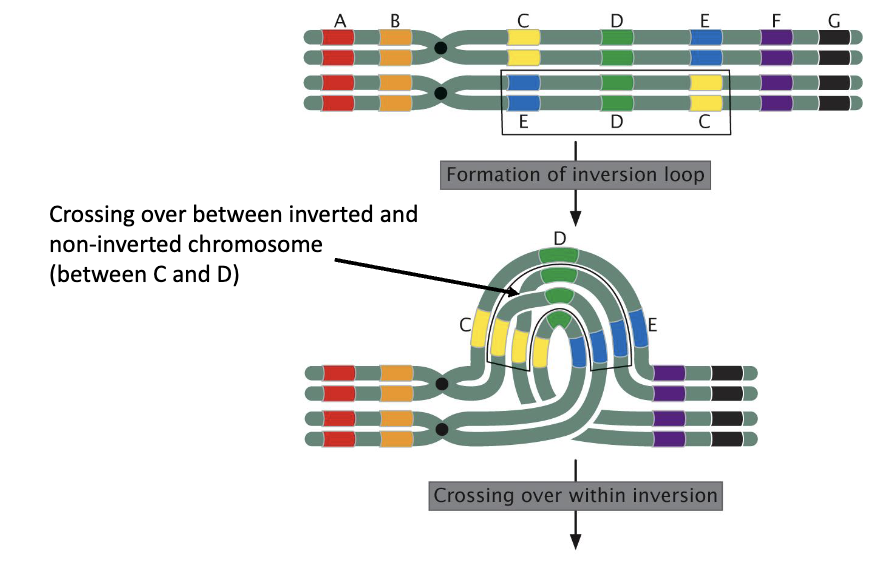
Dicentric Bridge
a physical linkage of two centromeres (one from the inverted chromosome, one from the normal) that forms during meiosis when crossing over occurs within the inversion loop
1this bridge gets stretched and broken as chromosomes separate, creating unbalanced gametes with deleted/duplicated segments and an acentric fragment
Crossing Over Within a Pericentric Inversion
crossing over between inverted and non-inverted chromosomes
C and D
results in reduced recombination frequency
reduced fertility
recombinant gametes are non viable because genes are missing or present in too many copies
Gamete Viability if No Crossing Over Occurs
if no crossing over occurs, gametes produced are usually viable because genetic information is not lost or gained
Gamete Viability if Crossing Over Occurs (OUTSIDE Inverted Region)
if crossing over occurs OUTSIDE of the inverted region, the gametes are viable.
Gamete Viability if Crossing Over Occurs (INSIDE Inverted Region)
if crossing over occurs INSIDE of the inverted region, some nonviable gametes and reduced recombination frequency
Translocation
exchange of segments between non-homologous chromosomes, or to a different region on the same chromosome
translocations between chromosomes can be reciprocal (two way) or non-reciprocal (one way)
if no genetic material is lost, considered a balanced translocation
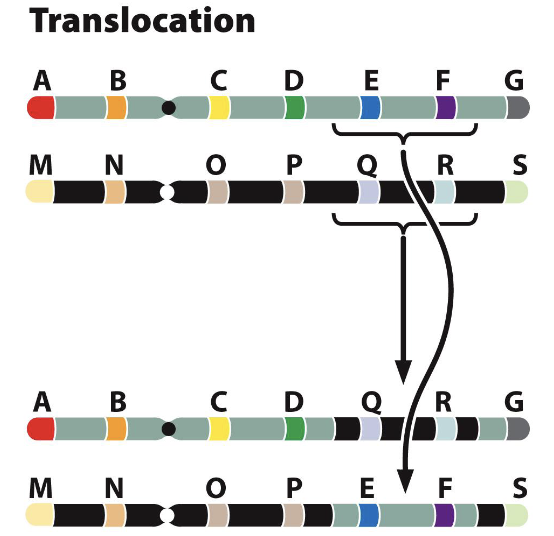
Reciprocal Translocations Consequences
as with inversions, translocations change the positions of genes
this can alter expression of genes because of association with different proteins, or formation of new gene products
fusion proteins
ex. philadelphia chromosomes
fused BCR-ABL genes
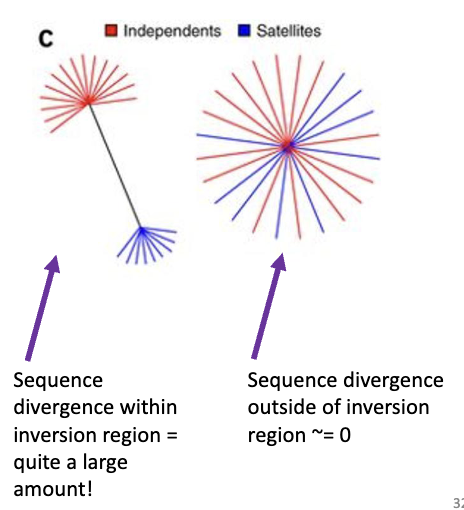
Inversions and Recombination
inversions suppress recombination
Lack of recombination within inversions means that genes within the inversions are free to diverge to produce different adaptations.
ex. Ruff bird
genes within alternate orientations of inversions can diverge dramatically even though there is no divergence anywhere else in the genome
no recombination within inversion
inside inversion: large divergence
outside inversion: no divergence
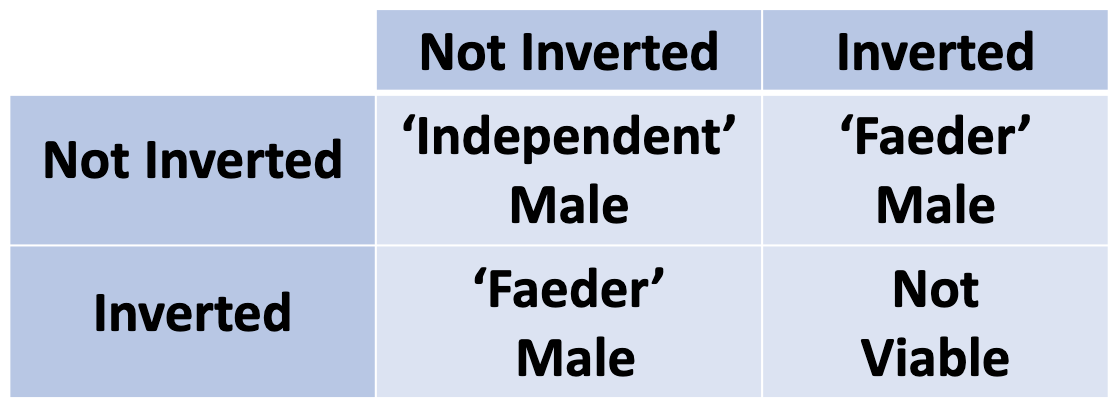
Ruff Bird Inversion Example
3 types of males: independent, faeder, satellite.
both faeder and satellite males have a genetic variation which arose nearly 4 million years ago
the inversion is lethal in the homozygous condition
Chromosomal Rearrangements in Atlantic Cod
cod have a large chromosomal inversion
genes inside the inversion influence whether cod are adapted to warmer or colder water
because recombination inside the inversion is suppressed, the warm and cold versions of the gene do not get scrambled by recombination
Terminology of Chromosome Types
Metacentric
centromere near the middle; arms are ~equal length.
Submetacentric
centromere slightly off-center; one arm longer.
Acrocentric
centromere very close to one end
Telocentric
centromere at the very end; essentially one arm (common in some animals, not humans
How Does Each Type of Chromosomal Variation Occur
Deletion: where a segment of a chromosome is lost
Chromosome breaks and the fragment is not reattached
Unequal crossing over during meiosis
DNA damage not properly repaired
Duplication: A chromosome segment is repeated.
Unequal crossing over during meiosis when homologous chromosomes misalign
Replication errors
Inversion: A chromosome segment breaks, flips, and reinserts.
Two breaks occur in one chromosome
Segment rotates 180° before rejoining
Paracentric – does not include the centromere
Pericentric – includes the centromere
Translocation: A segment moves to a non-homologous chromosome.
Chromosomes break and reattach incorrectly
Reciprocal translocation – two chromosomes exchange segments
Robertsonian translocation – two acrocentric chromosomes fuse at the centromere
Consequences of Each Chromosomal Variation
deletion: Loss of genes → often severe effects
Example: Cri-du-chat syndrome (5p deletion)duplication: Extra gene copies → altered gene dosage
Example: Bar eye mutation in Drosophilainversion: Usually no gene loss, but can disrupt meiosis and recombination
translocation: Often balanced in carriers, but can cause abnormal gametes. Example: Some cases of Down syndrome
Aneuploidy
a genetic condition where cells have an abnormal number of chromosomes, meaning they have extra or missing ones, deviating from the typical 46 in humans (23 pairs)
“somy” refers to the number of particular chromosomes
plants tolerate it better than animals
usually viable, though phenotype may be altered and fertility reduced
Aneuploidy in Human Genes
normal human diploid individual: 2n= 46
47 = (2n + 1)
gain of a single chromosome
48 = (2n + 2)
gain of a pair of homologous chromosomes
Trisomy
gain of a single chromosome
ex. 47 = (2n + 1)
Monosomy
loss of a single chromosome
ex. 45 = 2n - 1
Nullisomy
loss of both members of a pair of homologous chromosomes
ex. 44 = 2n - 2
Double Monosomic
loss of two members of non-homologous chromosomes
less common than nullisomy
ex. 44 = 2n - 2
Tetrasomy
gain of two homologous chromosomes
ex. 2n + 2 = 48
Double Trisomic
gain of two non-homologous chromosomes
ex. 2n + 2 = 48
Aneuploidy Origins
nondisjunction in meiosis or mitosis
deletion of a centromere leads to chromosome loss
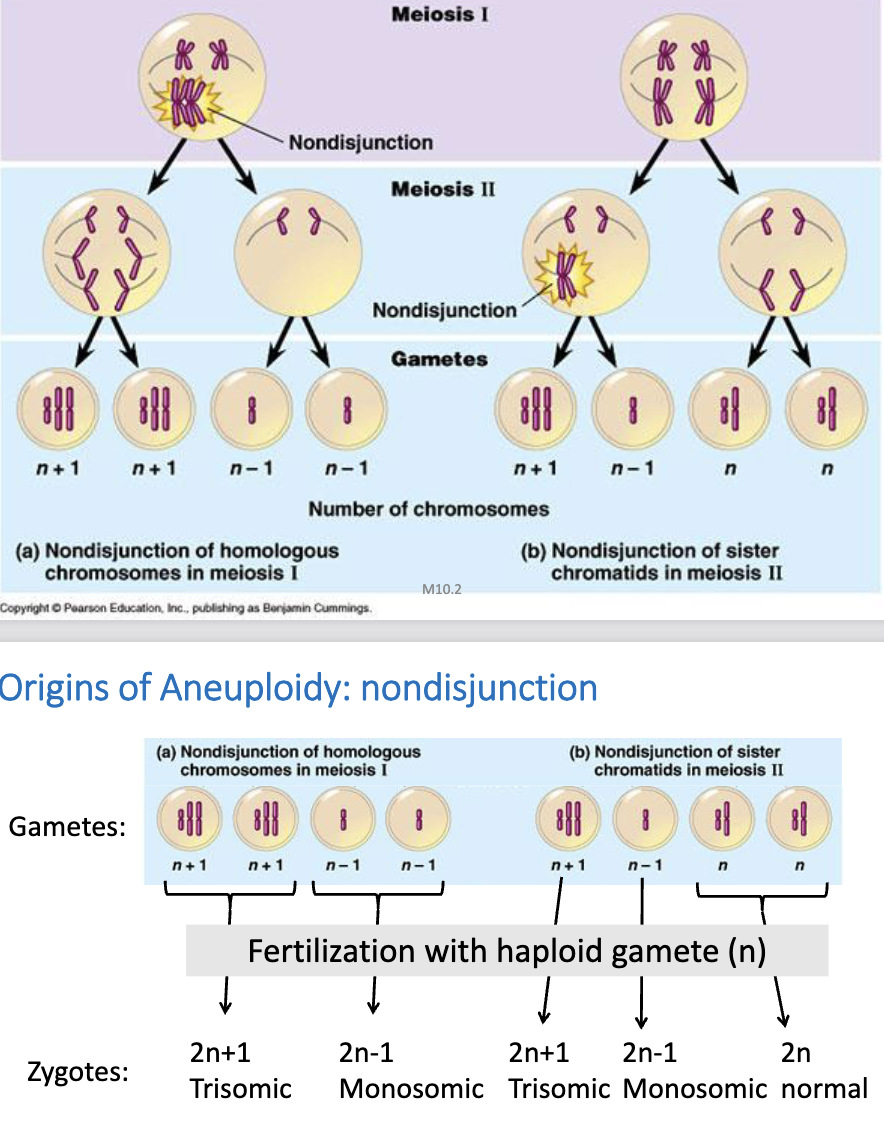
Nondisjunction Origin of Aneuploidy
failure of homologous chromosomes or sister chromatids to separate
may result in:
trisomic (still viable)
autosomal trisomy is thought to be the most common cause of miscarriages
monosomic (not viable, unless for sex chromosomes)
normal (still viable)
Trisomy 13
patau syndrome
viable
Trisomy 18
edwards syndrome
viable
Trisomy 21
down syndrome
viable
Monosomy X0
turner syndrome
only applies to girls
Primary Down Syndrome
trisomy 21
3 copies of chromosome 21
2n+1 = 47 chromsomes
accounts for most cases of Down Syndrome
most cases arise from random nondisjunction during meiotic division
mother contributes the extra chromosome in most cases
the incidence of trisomy 21 rises sharply with increasing maternal age
Familial Down Syndrome
an extra copy of chromosome 21 is attached to another chromosome, therefore cause of Down Syndrome in this case is Robertsonian translocation, not trisomy 21
accounts for 3-4% of cases
carrier parent: Has 45 chromosomes, not 46
One chromosome is a fusion of 15 + 21
Has normal phenotype (NOT Down syndrome)
depending on how those chromosomes segregate during meiosis, fertilisation can produce:
a normal child (carrier)
a child with Down syndrome
a non-viable embryo.
Trisomy 9
developmental delay and intellectual disability
physical abornmalities
musculoskeletal problems
gastrointestinal issues
however, trisomy 9 individuals survive because it is present as a mosaic, so not all cells have it.
Polyploidy
an increase in the number of sets of chromosomes
“ploidy” refers to the total number of chromosomes
common (and important) in plants, less common in animals, and not known at all in mammals (presumed to be lethal)
may be either:
autopolyploid
allopolypoid
Autopolyploid
multiples of the same genome
origin can occur during either mitosis or meiosis
nondisjunction of all chromosomes during mitosis in early embryo can produce autotetraploid
nondisjunction of all chromosomes during meiosis produces diploid games
Effects of Autopolyploidy
usually sterile
most gametes produces are genetically unbalanced
Autotriploid
diploid gamete + normal gamete
3n
Autotetraploid
diploid gamete + diploid gamete
4n
Allopolyploid
multiples of closely related genomes
How to Solve Infertility
to convert sterile hybrid into fertile new species, they need chromosome doubling
hybrid is sterile
unbalanced gametes are nonviable
but if ehte entire genome is doubled by mitotic non-disjunction, the fertility problem is solved
Polyploids in Agriculture
cell volume correlated with nucleus volume, correlated with genome size
polyploids often have bigger leaves, fruits, and seeds
bread wheat is a polyploid derived from 3 species
Mutations
mutations are both rare and common
rare: because DNA with replication occurs with high fidelity
common: because there is a lot of DNA being replicated
there is about 64 new mutations in each human generation
classified based on transmission
somatic
germ-line
Somatic Mutations
are not transmitted from one generation to another
Germ-line Mutation
may be transmitted to 50% of offspring
Point Mutations Classification
classified by their effect on amino acid sequence of proteins
can be classidfied into 3 categories, based on effect:
silent (synonymous): no change in amino acid sequence
missense (nonsynonymous): causes 1 aa to be substituted for another
nonsense: aa codon is substituted for stop condo
Indels
cause frameshifts that alter reading frames, creating either nonsense or missense effects on protein
except when indels occur as multiples of 3 nucleotides. in such cases, the amino acid sequences will change, but the reading frame is preserved
indels outside of reading frames usually have no effect on phenotype
Loss-of-function Mutation
protein function completely or partially lost
recessive inheritance
Gain-of-function Mutation
new gene product, or gene product in the wrong tissue
dominant inheritance
Neutral Mutation
missense mutation that results in non-significant change in protein function, because one chemically similar amino acid substituted for another, or occurs in a part of the protein that is not important for function
Transition Point Mutation
a common DNA change where a purine (Adenine/A or Guanine/G) gets swapped for another purine, or a pyrimidine (Cytosine/C or Thymine/T) is swapped for another pyrimidine, maintaining the base's ring structure class (e.g., A→G, C→T)
Transversion Point Mutation
a genetic change where a purine (Adenine or Guanine) is swapped for a pyrimidine (Cytosine or Thymine), or vice versa, altering the DNA's ring structure, unlike transitions
Forward Mutation
alters the wild phenotype
Reverse Mutations
changes mutant phenotype back to wild phenotype.
true reversions
suppressor mutations
Suppressor Mutation
where the first mutation is suppressed by a second mutation
intragenic suppressor mutation
intergenic suppressor mutation
Intragenic Suppressor Mutation
in the same gene
intra = “within”
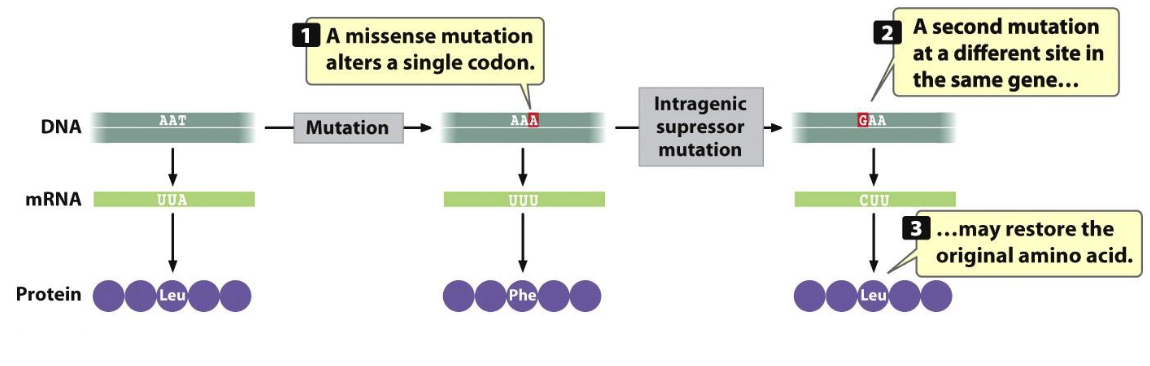
Intergenic Suppressor Mutation
a second mutation that occurs in a different gene from the original mutation, and it counteracts the original mutation's effects to restore the normal phenotype
inter = “between”
How do Mutations Occur?
spontaenously
induced by physical and chemical agent
Spontaneous Mutations
caused by errors during DNA replication
tautomeric shift
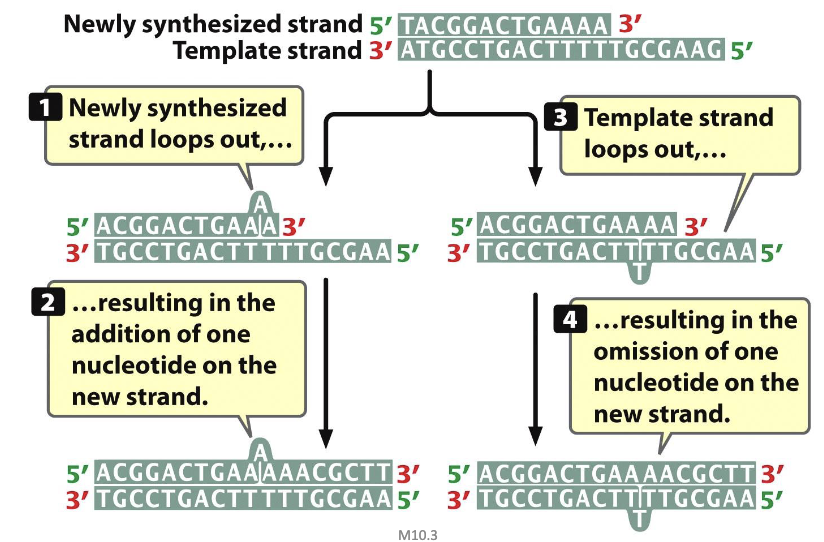
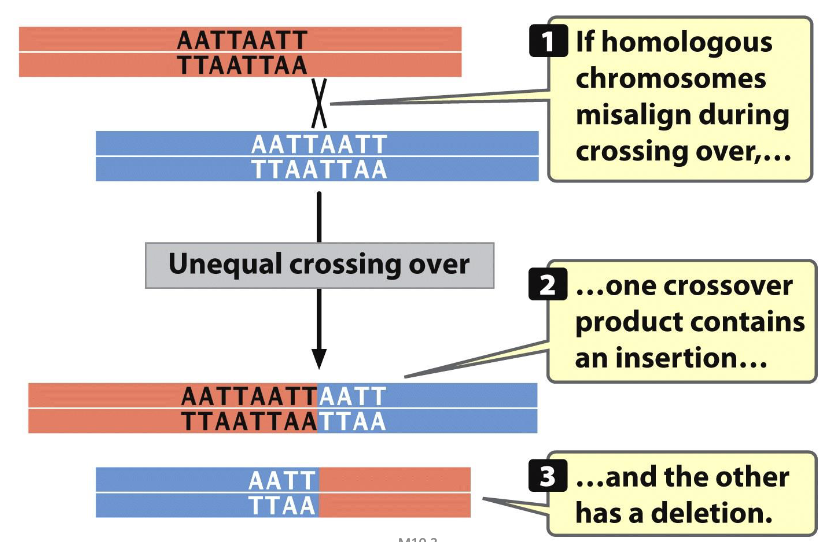
DNA strand slippage
misalignment
Tautomeric Shifts (spontaneous mutations)
base tautomers can incorrect base-pair during DNA replication, thus creating a transition mutation
DNA Strand Slippage (spontaneous mutations)
insertion/deletion owing to slipped-strand mispairing during DNA replication
Misalignment (spontaneous mutations)
misalingment of homologous chromosomes during crossing-over at meiosis I
Mutagen Agents that Cause Mutation
radiation
chemical:
base analogs
base modifying agents
intercalating agents
Radiation Mutagen Types
ionizing
ultraviolet
Ionizing Radiation
creates free radicals
cosmic, gamma, or xrays bombard an atom, which dislodges an electron, and results in a change in stable molecules into a free radical which can alter the structure of bases and break phosphodiester bonds in DNA
Ultraviolet Radiation
electromagnetic radiation of lower energy than ionizing radiation
can still generate free radicals under some circumstances, but less likely to do some than higher energy radiation
most common source is the sun
though can be generated by various types of lamps
Nucleotide Excision Repair
dna repair enzymes correct damaged DNA…
protein recognizes mismatches
unwinds DNA in area of mismatch
excises out nucleoides
fills in correct nucleotides
Xeroderma Pigmentosum
an autosomal recessive genetic disorder of DNA repair
the ability to repair mutations caused by UV light is deficient
Chemical Mutagens Type
base analogs
base modifying agents
intercalating agents
Base Analogs
chemicals that appear similar to the normal bases in DNA, but causes incorrect base-pairing and introduce point mutations during DNA replication
ex. 5-Bromouracil: a nucleotide analong which resembles both thymine and cytosine (when ionized, it pairs with guanine rather than adenine)
Base Modifying Agents
chemicals that modify groups on the normal bases in DNA that result in incorrect base-pairing and introduce point mutations during DNA replication
Intercalating Agents
intercalating agents insert between adjacent bases, distorting them by 0.68 nm, the size of a base
first round of DNA replication, the DNA polymerase randomly selects any nucleoside triphosphate opposite the intercalating agent
result: frame-shift due to insertion of a base
Ames Test
assay for chemical mutagenicity via His- Salmonella
His- Salmonella cannot grow on minimal medium lacking the essential amino acid, histidine
His+ Salmonella will grow on the minimal medium
increased reversions of His- to His+ Salmonella indicates the chemical is a mutagen, and thus a potential carcinogen
How Does the Ames Test Work?