Ic50
1/62
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
63 Terms
What is an enzyme?
A protein
What do enzymes do?
They catalyse the conversion of substrates (S) to products (P).
Are enzymes chemically unchanged or changed at the end of a reaction?
Unchanged
Do enzymes change the equilibrium position of the reaction?
No
Enzyme catalytic cycles
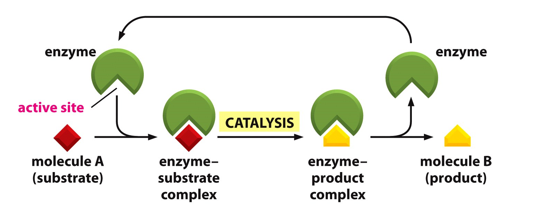
What are the 2 models of catalysis?
Lock and key
Induced Fit
Thermodynamic catalysis by enzymes
Most enzymes work by lowering activation energy of the reaction

What is the chemical catalysis of enzyme reactions?
The process by which enzymes speed up (catalyse) chemical reactions in the body by using chemical interactions at their active site.
How do enzymes increase the likelihood of substrate molecules reacting?
By holding substrates and reacting groups close together in the active site.
Why is orientation of reacting groups important in enzyme reactions?
Correct orientation ensures bonds react efficiently, lowering activation energy.
How do enzymes mimic the transition state?
By stretching or bending reacting bonds, making them easier to break.
Why are enzyme active sites often hydrophobic?
To exclude water, stabilize charged groups, and create a favorable reaction environment.
What is the effect of excluding bulk solvent from enzyme active sites?
Charged groups are not masked, enhancing electrostatic interactions.
How can water molecules participate in enzyme-catalyzed reactions?
Ordered water molecules can act as reactants or help stabilize intermediates.
How do enzymes perform acid/base catalysis?
Active site groups donate or accept protons to facilitate bond breaking/forming.
What is nucleophilic catalysis in enzyme reactions?
Formation of a temporary covalent bond (e.g., acyl-enzyme intermediate) with the substrate.
Why are enzymes usually stereospecific?
Their chiral active sites can only accommodate substrates of a certain orientation.
How do cofactors and cosubstrates help enzyme activity?
Metal ions or nucleotide-derived molecules assist in catalysis and are often derived from vitamins/minerals.
How do enzymes stabilize the transition state?
Active sites are complementary to the transition state, distorting bonds to lower activation energy.
Chemical model of Enzyme Catalysis:
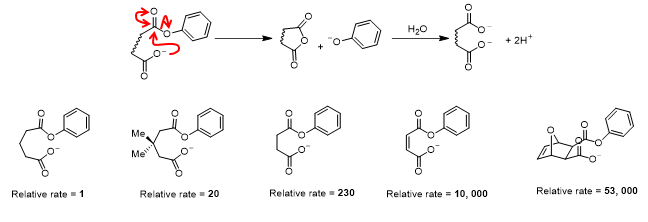
Exam Tip
Small molecules should have the correct bond angles, i.e. a sp3 carbon is tetrahedral (109) and a sp2 carbon is trigonal planar (120)
Non reacting parts of the molecule can be designated as ‘R’
Only necessary to draw enzyme residue side chains when covalent interme
Acyl substitution reactions
Activation of active site nucleophile by deprotonation
Acyl substitution using active site nucleophile
Acyl substitution by water to regenerate enzyme
In acyl substitution reactions, what do poor leaving groups require?
Protonation
Do acyl substitution reactions occur for aldehydes and ketones?
No because H atoms, alkyl groups, and aryl groups cannot leave
Step 1) Chymotrypsin
Activation of nucleophile
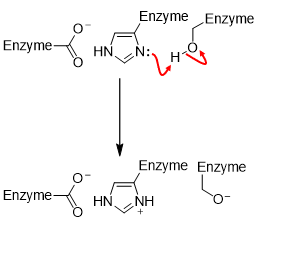
Step 2) Chymotrypsin. - Formation of an acyl-enzyme intermediate
Deprotonated nucleophile adds to carbonyl group.
Tetrahedral intermediate is transiently formed.
Leaving group is protonated (-NH2 produced).
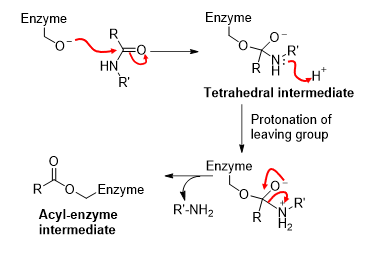
Step 3) Chymotrypsin - Hydrolysis of acyl-enzyme intermediate
Mechanism is similar, with formation of tetrahedral intermediate. This time, water is the nucleophile and the enzyme-O- is the leaving group.
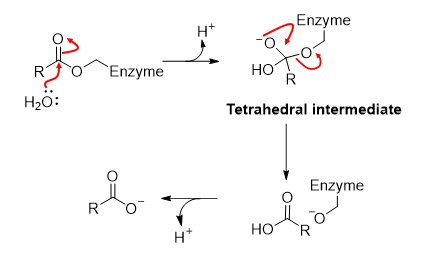
Name all the common ways enzymes catalyse reactions:
Entropic effects.
Bond stretching and bending.
Acid / base catalysis.
Nucleophilic catalysis.
Removal of bulk solvent and use of ordered waters.
Use of cofactors and cosubstrates.
Name the factors that affect enzyme activity:
pH – due to changing ionisation state of side-chains. Grossly inappropriate pH unfolds proteins;
Denaturing reagents (detergents, urea, guanidinium hydrochloride);
Temperature – higher temperatures increase rate until thermal denaturation (unfolding) of the protein occurs
Activity is proportional to amount of enzyme
Substrate concentration
The presence of inhibitors
How do enzymes work?
Enzymes are catalysts, therefore enzyme concentration is usually far greater than substrate concentration
The reaction goes through an enzyme/substrate complex
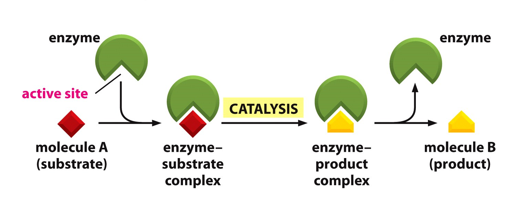
What do simple models assume?
Only one molecule of one substrate binds to the enzyme (One substrate reactions are rare – isomerizations and rearrangements);
Enzyme and substrate form a [ES] complex (rate-limiting step);
Enzyme converts substrate to product and product release occurs (fast);
Products bind weakly to enzymes.
Name the 3 simple models:
Differences in simple models:
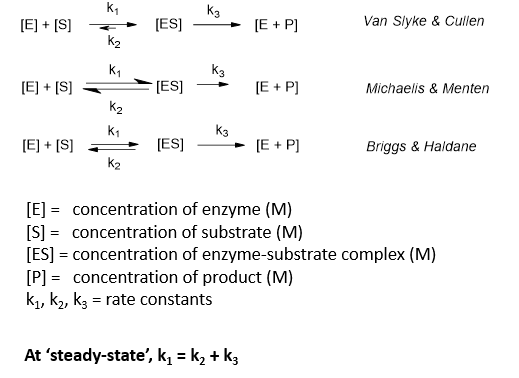
Michaelis Menten curve
v = rate;
[S] = substrate concentration;
Vmax = rate when all enzyme active sites are occupied (the enzyme is saturated);
Km = [S] at which v = ½ Vmax
![<ul><li><p><span><em><span>v</span></em><span> = rate;</span></span></p></li><li><p style="text-align: justify;"><span><span>[S] = substrate concentration;</span></span></p></li><li><p style="text-align: justify;"><span><em><span>V</span></em><sub><span>max</span></sub><span> = rate when all enzyme active sites are occupied (</span><em><span>the enzyme is saturated</span></em><span>);</span></span></p></li><li><p style="text-align: justify;"><span><em><span>K</span></em><sub><span>m </span></sub><span>= [S] at which </span><em><span>v</span></em><span> = ½ </span><em><span>V</span></em><sub><span>max</span></sub></span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/600db4d2-768b-495e-9ea7-2e8dea6c0f76.png)
What’s the Michaelis-Menten equation?
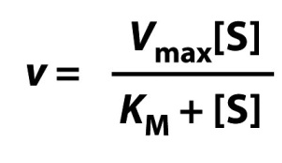
Why do enzyme reactions saturate?
Enzyme reactions saturate because there are a limited number of enzyme active sites available to bind substrate molecules.
At low substrate concentrations, increasing the substrate makes the reaction go faster — because more enzyme–substrate complexes can form.
However, as substrate concentration keeps increasing, all enzyme active sites eventually become occupied.
Explaining why enzyme reactions saturate using the Michaelis-Menten equation:
Rearrange ‘Michaelis-Menten’ equation: (v/Vmax)= ([S]/(Km +[S] ))
Assume Km = 1 mM:
At 1 mM S, v = 0.5 Vmax At 2 mM S, v = 0.666 Vmax
At low [S] rate (v) increases with increasing [S] (first order);
At 20 mM S, v = 0.952 Vmax At 21 mM S, v = 0.954 Vmax
At high [S], rate (v) does not increase with further increases in [S] (zero order). Rate only depends on [ES] and k3 (i.e., saturation has occurred).
![<ul><li><p><span><span>Rearrange ‘Michaelis-Menten’ equation: (</span></span><span style="font-family: "Cambria Math";"><span>v/Vmax)= ([S]/(</span></span><span><strong><span>K</span><sub><span>m</span></sub></strong></span><span style="font-family: "Cambria Math";"><strong><sub><span> </span></sub></strong><span>+[S] ))</span></span></p></li><li><p style="text-align: justify;"><span><strong><span>Assume </span><em><span>K</span></em><sub><span>m</span></sub><span> = 1 mM:</span></strong></span></p></li><li><p style="text-align: justify;"><span><span>At 1 mM S, </span><em><span>v</span></em><span> = 0.5 </span><em><span>V</span></em><sub><span>max </span></sub><span>At 2 mM S, </span><em><span>v</span></em><span> = 0.666 </span><em><span>V</span></em><sub><span>max </span></sub></span></p></li><li><p style="text-align: justify;"><span><span>At low [S] rate (</span><em><span>v</span></em><span>) increases with increasing [S] (</span><strong><span>first order</span></strong><span>);</span></span></p></li><li><p style="text-align: justify;"><span><span>At 20 mM S, </span><em><span>v</span></em><span> = 0.952 </span><em><span>V</span></em><sub><span>max </span></sub><span>At 21 mM S, </span><em><span>v</span></em><span> = 0.954 </span><em><span>V</span></em><sub><span>max</span></sub></span></p></li><li><p style="text-align: justify;"><span><span>At high [S], rate (</span><em><span>v</span></em><span>) does not increase with further increases in [S] (</span><strong><span>zero order</span></strong><span>). Rate only depends on [ES] and k</span><sub><span>3 </span></sub><span>(</span><em><span>i.e.,</span></em><span> saturation has occurred).</span></span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/f4ca78e6-88a4-4b1e-ac5d-0f6dc8ec68ec.png)
What are the assumptions of the steady state model?
Enzyme concentration is much smaller than substrate concentration ([E] « [S]) (typically nM vs. mM);
[S] remains constant; Measured rate of reaction is linear at beginning of curve (‘initial rate’);
[ES] remains the same throughout course of reaction (‘The steady-state approximation’);
Product binds weakly to enzyme;
Rate of conversion of product to substrate is negligible (no backward reaction)-
Michaelis-Menten Plot
Directly plots v against [S]
Can be difficult to decide when Vmax is reached
Usually requires a computer programme
![<ul><li><p>Directly plots v against [S]</p></li><li><p>Can be difficult to decide when V<sub>max</sub> is reached</p></li><li><p>Usually requires a computer programme</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/f6bec42e-14f1-45d8-a7f1-25ab572782fb.png)
Lineweaver-Burk Plot
Plots 1/v against 1/[S]
Y-intercept = 1/Vmax
X-intercept = -1/Km
Higher precision;
Lower accuracy;
Errors are not equal at all points (least squares regression is not appropriate).
![<ul><li><p>Plots 1/v against 1/[S]</p></li><li><p><span><span>Y-intercept = 1/</span><em><span>V</span></em><sub><span>max</span></sub></span></p></li><li><p><span><span>X-intercept = -1/</span><em><span>K</span></em><sub><span>m</span></sub></span></p></li><li><p><span><span>Higher precision;</span></span></p></li><li><p><span><span>Lower accuracy;</span></span></p></li><li><p><span><span>Errors are not equal at all points (least squares regression is </span><strong><span>not </span></strong><span>appropriate).</span></span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/1d614257-b351-4fde-8f94-a94d4cc565c9.png)
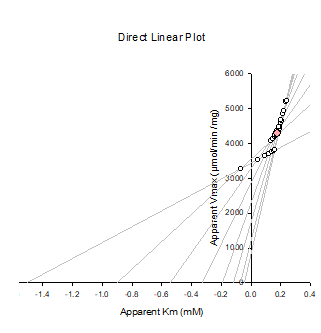
Direct Linear Plot
Plot rate and substrate concentration onto axes;
Connect points;
Intersections are estimates of Km and Vmax;
Gives median value;
Makes no assumptions about the errors;
Difficult to deviations from ideal behaviour.
Vmax
The fastest rate the enzyme can work at- when all the enzyme active sites are full of substrates (all enzymes are saturated)
If Vmax = 0.5 what does that mean?
The maximum rate of the enzyme catalysed reaction is 0.5 units per second. When the enzyme is working as fast as it possibly can (all enzyme active sites are full of substrate), it can make 0.5 units of product per unit time.
Km
It tells you how strongly an enzyme binds to its substrate.
A low Km means the enzyme binds tightly (it works well even at low substrate levels).
A high Km means the enzyme binds weakly (it needs more substrate to work efficiently).
What is Km often similar to?
Substrate concentration in cell
Kcat
Kcat shows how fast the enzyme can convert substrate to product once the substrate is bound.
Vmax / amount of enzyme in, for example, nmoles.
This is the first order rate constant for conversion of substrate to product.
Kcat = K3
Exam Question: In an enzyme kinetic experiment, the value for 1/Vmax was 0.102 min/nmol and -1/Km was -3.45 mM-1 from a Lineweaver-Burk (double reciprocal) plot. Each assay contained 0.14 mg of enzyme with a molecular weight (MW) of 47 146 Da. Showing your workings, calculate values for Vmax, Km, kcat, kcat/Km, assuming one active site per monomer (hint: kcat = Vmax divided by amount of enzyme) [10 marks].
Ki
Inhibition constant
Measures how strongly an inhibitor binds to the enzyme
Ki values related to binding energy (ΔG)
A low Ki means the inhibitor is very effective (binds strongly - more potent).
A high Ki means it’s a weaker inhibitor (binds less tightly - less potent).
Mode of action for Inhibitors
Inhibitors can resemble the substrate in structure
Inhibitors affect either Km, Vmax or both
Competitive inhibition
Km increases. X-intercept changes
Km(I) = Km (1 + [I]/Ki)
Km(I) = apparent Km (with inhibitor)
[I] = inhibitor concentration
Ki = inhibition constant (strength of inhibitor)
No change in Vmax. Y-intercept does not change
Inhibition decreases with increased substrate concentration
![<ul><li><p>K<sub>m</sub> increases. X-intercept changes</p></li><li><p><span><strong><em><span>K</span></em><sub><span>m</span></sub><span>(I) = </span><em><span>K</span></em><sub><span>m</span></sub><span> (1 + [I]/</span><em><span>K</span></em><sub><span>i</span></sub><span>)</span></strong></span></p></li></ul><p><strong><em>K</em><sub>m</sub>(I)</strong> = apparent Km (with inhibitor)</p><p><strong>[I]</strong> = inhibitor concentration</p><p><strong><em>K</em><sub>i</sub></strong> = inhibition constant (strength of inhibitor)</p><ul><li><p>No change in V<sub>max</sub>. Y-intercept does not change</p></li><li><p>Inhibition decreases with increased substrate concentration</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/d571bc2a-1822-4908-be35-20e071c52cf8.png)
Non-competitive inhibition
Vmax is reduced. Y-intercept changes.
Vmax/Km (I) = Vmax/Km(1 + [I]/Ki)
Vmax = The enzyme’s maximum velocity — how fast it can go when fully saturated with substrate.
Km (I) = apparent Km (with inhibitor)
[I] = inhibitor concentration
Ki = inhibition constant (strength of inhibitor)Km is not affected. X-intercept does not change.
Inhibition is not reduced by increasing substrate.
![<ul><li><p><span><em><span>V</span></em><sub><span>max</span></sub><span> is reduced. Y-intercept changes.</span></span></p></li><li><p style="text-align: justify;"><span><strong><em><span>V</span></em><sub><span>max</span></sub><span>/</span><em><span>K</span></em><sub><span>m</span></sub><span> (I) = </span><em><span>V</span></em><sub><span>max</span></sub><span>/</span><em><span>K</span></em><sub><span>m</span></sub><span>(1 + [I]/</span><em><span>K</span></em><sub><span>i</span></sub><span>)</span></strong></span></p></li></ul><p style="text-align: justify;"><strong><span>V</span><sub><span>max</span></sub></strong><sub><span></span></sub> = The enzyme’s maximum velocity — how fast it can go when fully saturated with substrate.</p><p style="text-align: justify;"><strong><em>K</em><sub>m </sub>(I)</strong> = apparent Km (with inhibitor)</p><p><strong>[I]</strong> = inhibitor concentration</p><p><strong><em>K</em><sub>i</sub></strong> = inhibition constant (strength of inhibitor)<span><em><span>K</span></em><sub><span>m</span></sub><span> is not affected. X-intercept does not change.</span></span></p><ul><li><p style="text-align: justify;"><span><span>Inhibition is </span><strong><span>not </span></strong><span>reduced by increasing substrate.</span></span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/df62561b-5c32-4375-8500-adbeacb810b1.png)
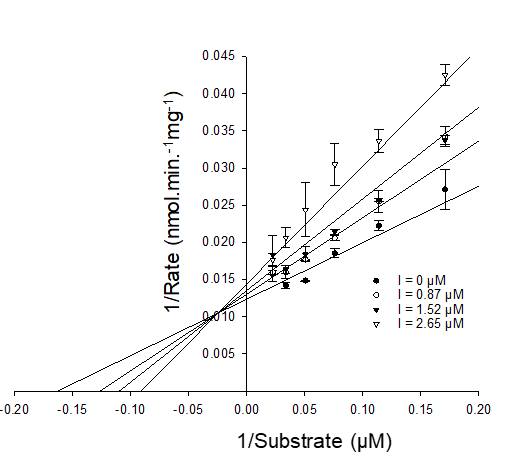
Mixed-competitive inhibition
Km is increased (as for competitive inhibition). X-intercept changes.
Vmax is decreased (as for non-competitive inhibition). Y-intercept changes.
α factor defines the competitive and non-competitive components.
Uncompetitive inhibition
Inhibitor binds to the ES complex
Parallel lines in Lineweaver-Burk plot
Both Km and Vmax reduced by same amount, (1 + [I]/Ki)
Increasing substrate concentration increases inhibition.
![<ul><li><p style="text-align: justify;"><span><span>Inhibitor binds to the ES complex</span></span></p></li></ul><ul><li><p style="text-align: justify;"><span><span>Parallel lines in Lineweaver-Burk plot</span></span></p></li><li><p style="text-align: justify;"><span><span>Both </span><em><span>K</span></em><sub><span>m</span></sub><span> and </span><em><span>V</span></em><sub><span>max</span></sub><span> reduced by same amount, (1 + [I]/</span><em><span>K</span></em><sub><span>i</span></sub><span>)</span></span></p></li><li><p style="text-align: justify;"><span><strong><span>Increasing</span></strong><span> substrate concentration </span><strong><span>increases </span></strong><span>inhibition.</span></span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/6ca8a47c-efc6-45ef-91e2-08ae874c4f1b.png)
What do dose response curves measure?
They measure the drug potency at a fixed substrate concentration

Example of a dose response curve:
IC50 curves
IC50 curves
IC50 = concentration of drug required for 50% reduction in activity
Value depends on assay conditions (especially substrate concentration)
IC50 not the same as Ki (can be calculated from IC50 for some inhibitors).
What is tight-binding inhibition?
Tight-binding inhibition means the inhibitor binds so strongly to the enzyme that it’s almost irreversible — the inhibitor and enzyme form a very stable complex.
Tight-binding inhibition means the inhibitor “grabs” the enzyme so strongly that it barely lets go. It’s like a super strong magnet — not a casual, reversible attachment.
These inhibitors are very potent and often used as drugs because they effectively “shut down” enzymes even at low concentrations.
When does tight binding inhibition occur?
When the concentrations of active enzyme and inhibitor are similar.
This means that the concentration of free inhibitor does not approximate to total inhibitor
In tight binding inhibition, what will happen to the Ki value if you increase the enzyme concentration?
It will increase
Features of tight-binding inhibition
The apparent Ki value will increase with increasing enzyme concentration;
Tight-binding inhibition often has very slow onset, and the enzyme inhibitor complex can have a very long half-life (Ki = koff/kon);
This means that infrequent dosing regimens can be used (easier for patient, reduced side-effects etc.). Therefore, many drug discovery programmes aim to develop a tight-binding inhibitor.
What’s the importance of Ki as the parameter defining potency?
Independent of enzyme and substrate concentrations
Unlike IC₅₀ (the inhibitor concentration that reduces activity by 50%), Ki is a true thermodynamic constant — it depends only on binding strength, not on experimental conditions.
This makes it a universal measure for comparing inhibitors across studies.
Directly reflects binding affinity
Ki comes from the equilibrium between enzyme and inhibitor — it quantifies how “sticky” the inhibitor is for the enzyme.
Useful for drug design
In pharmacology, a low Ki means the drug can be effective at low doses, reducing side effects and increasing specificity.
Irreversible Inhibitors
Examples include penicillins and other β-lactam antibiotics;
Enzymes inhibited are penicillin-binding proteins (PBPs);
Inhibitors act as pseudo-substrates but get ‘stuck’ (at acyl-enzyme intermediate);
Kinetic behaviour is more similar to chemical systems.
Exam Question) In an enzyme kinetic experiment -1/Km was -5.0 mM-1 and in the presence of an inhibitor -1/Km was -1.818 mM-1 when measured using a Lineweaver-Burk (double reciprocal) plot. The value for 1/Vmax did not change. In this experiment 25 μL of a 10 mM stock solution of inhibitor was added to a final assay volume of 1 mL. What sort of inhibitor is it? Show your workings and calculate the Ki value [9 marks]