MCB 250 – Replication Initiation and Telomerase
1/59
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
60 Terms
How many initiation sites does E. coli chromosome have?
one → oriC
Which protein is oriC recognized by?
a specific initiator protein, DnaA
Does DnaA require energy?
yes
What does the binding of DnaA-ATP do?
it begins a series of events that leads to the establishment of replication forks
How controlled is replication initation?
tightly controlled
How long is the oriC region?
~250 bps
What is the oriC region composed of?
multiple DnaA binding sites next to an A=T rich region
Which sequence does DnaA-ATP recognize?
the A=T rich rigion
What is DnaA-ATP and what does it do?
a nucleoprotein complex that causes unwinding of the DNA
How does DnaA-ATP open up the A=T rich region?
DNA binding around ATP increases negative supercoiling which opens the region up
What are the steps of replication initiation?
DnaA-ATP binds to its recognition sites in oriC
DnaA-ATP causes unwinding of the DNA
DnaC loads DnaB (helicase) onto the ssDNA
one helicase on each strand
helicase travels 5’ to 3’ and recruits primase → initiates DNA synthesis
Draw a diagram for replication initiation. Include origin(s), leading and lagging strands, directionality, and label the first lagging strand Okazaki fragment(s).
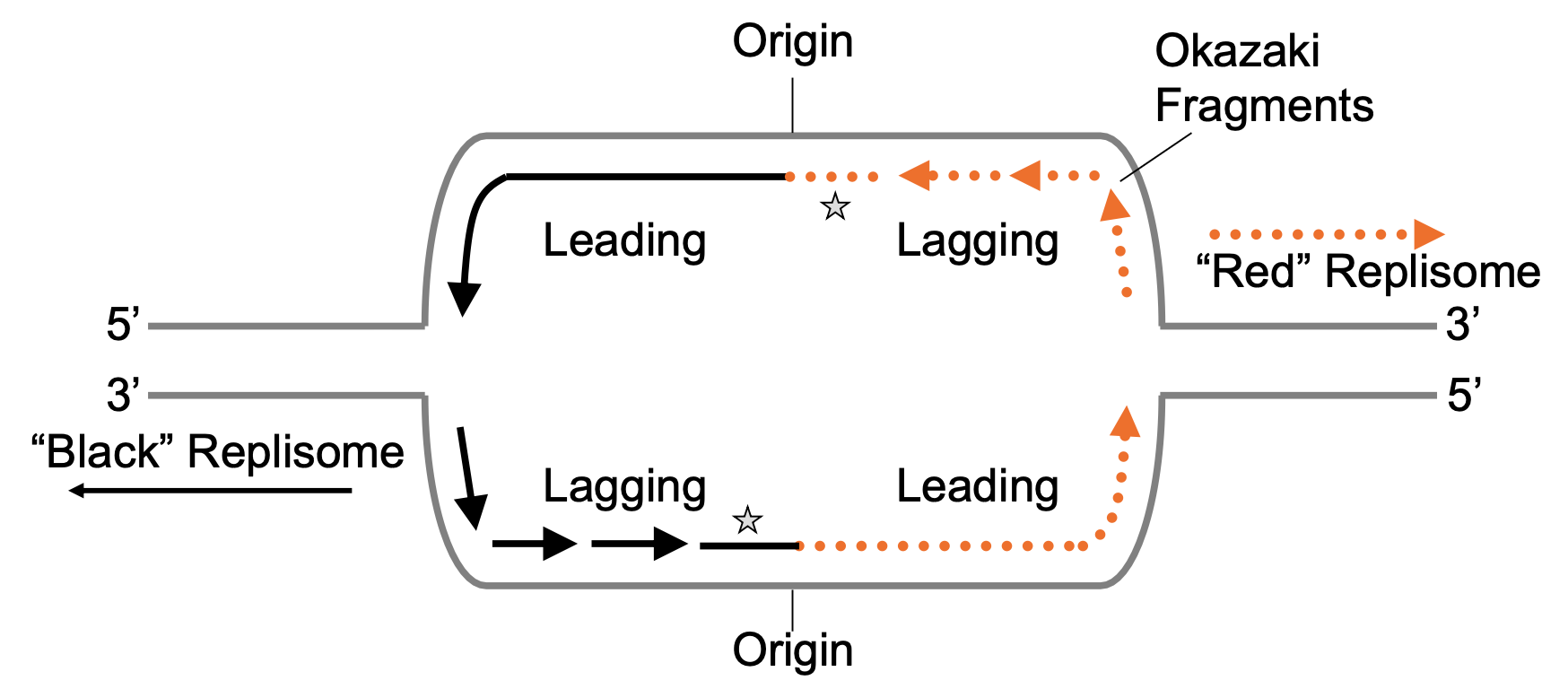
Where does replication initiation only need to occur?
only on the lagging strand → the first lagging strand can act as the leading strand for the other replisome
What does the replisome MUST be capable of?
re-initiating the leading strand
Is cell division and DNA replication coordinated?
yes
What must be coordinated with DNA replication?
cell division
Why does DNA replication and cell division have to be coordinated?
each daughter cell needs to get a fully replicated chromosome
How does DNA replication and cell division achieve coordination?
Is DNA in E. coli methylated?
yes
What is Dam?
DNA Adenine Methyltransferase
What does Dam do?
it recognizes the sequence 5’-GATC-3’ which is found 1/256 base pairs, and methylates the A’s in the major groove
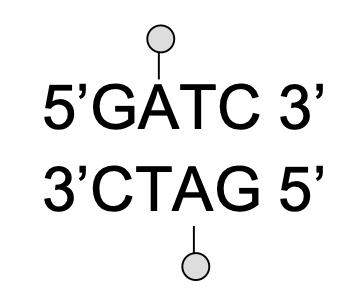
What does methylation tell us?
which strand is old DNA → is methylated
which strand is newly synthesized → is not methylated yet
Which strand – old or new – is methylated in DNA?
old strand
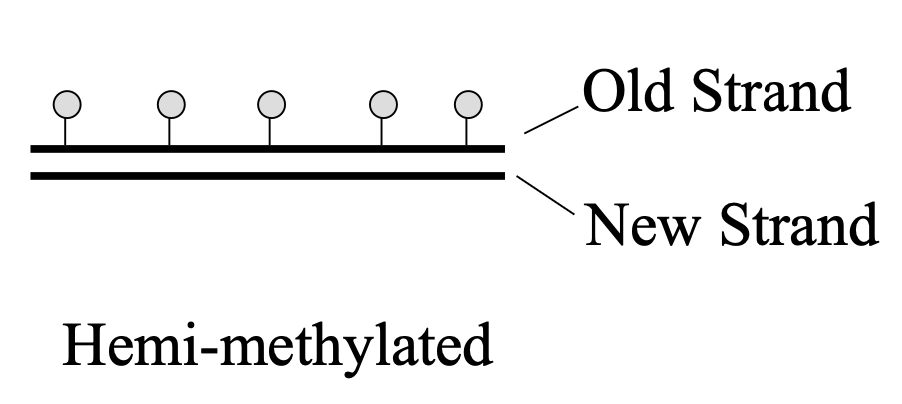
What is hemi-methylated DNA?
methylated old strand and non-methylated new strand
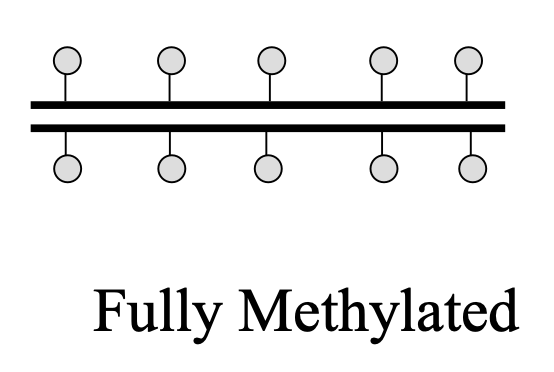
What is fully methylated DNA?
when both strands, old and new, are methylated
What is the speed of methylation in comparison to replication?
methylation is significantly slower than replication
How is replication initiation controlled?
control of DnaA-ATP levels
control of access to oriC
What is the rate-limiting step in replication initiation?
the binding of DnaA-ATP to oriC
How does DnaA-ATP regulate replication initiation?
the concentration of DnaA-ATP is tightly controlled
the amount of DnaA is proportional to cell mass ∴ small (new) cells don’t have enough DnaA to initiate replication
DnaA-ATP is converted to DnaA-ADP as the replication fork passes
DnaA-ADP doesn’t work as an initiator and takes a long time to convert back to DnaA-ATP ∴ slowing down replication initiation
How does the oriC regulate replication initiation?
oriC is inactive until it’s fully methylated
DnaA-ATP binds to fully methylated DNA
however, protein SeqA binds hemimethylated oriC (which is present before fully methylation) which prevents DnaA from binding ∴ slowing down Dam methylation and oriC activation
What does Seq A do?
it binds to hemimethylated oriC → prevents DnaA and Dam from binding → slows down re-initiation
How is replication terminated?
*not completely understood
the replisomes collide and everything gets unloaded
Pol I fills in any gaps
ligase seals any nicks
What does Topo IV do in replication?
it is required for recatenation
What are Type II topoisomerases used for in replication?
to relieve positive supercoils in front of the replication fork
gyrase does this in E. coli
to resolve catenanes
Topo IV does this in E. coli
Why does the replisome move more slowly in eukaryotes than in E. coli?
probably due to chromatin structure
How many origins of replication do eukaryotic chromosomes have?
multiple; perhaps thousands
Why do humans need more origins of replication than E. coli?
humans have 1000x more DNA than E. coli so replication needs to be initiated simultaneously at multiple origins on each chromosome
Draw the basic steps for eukaryotic replication.
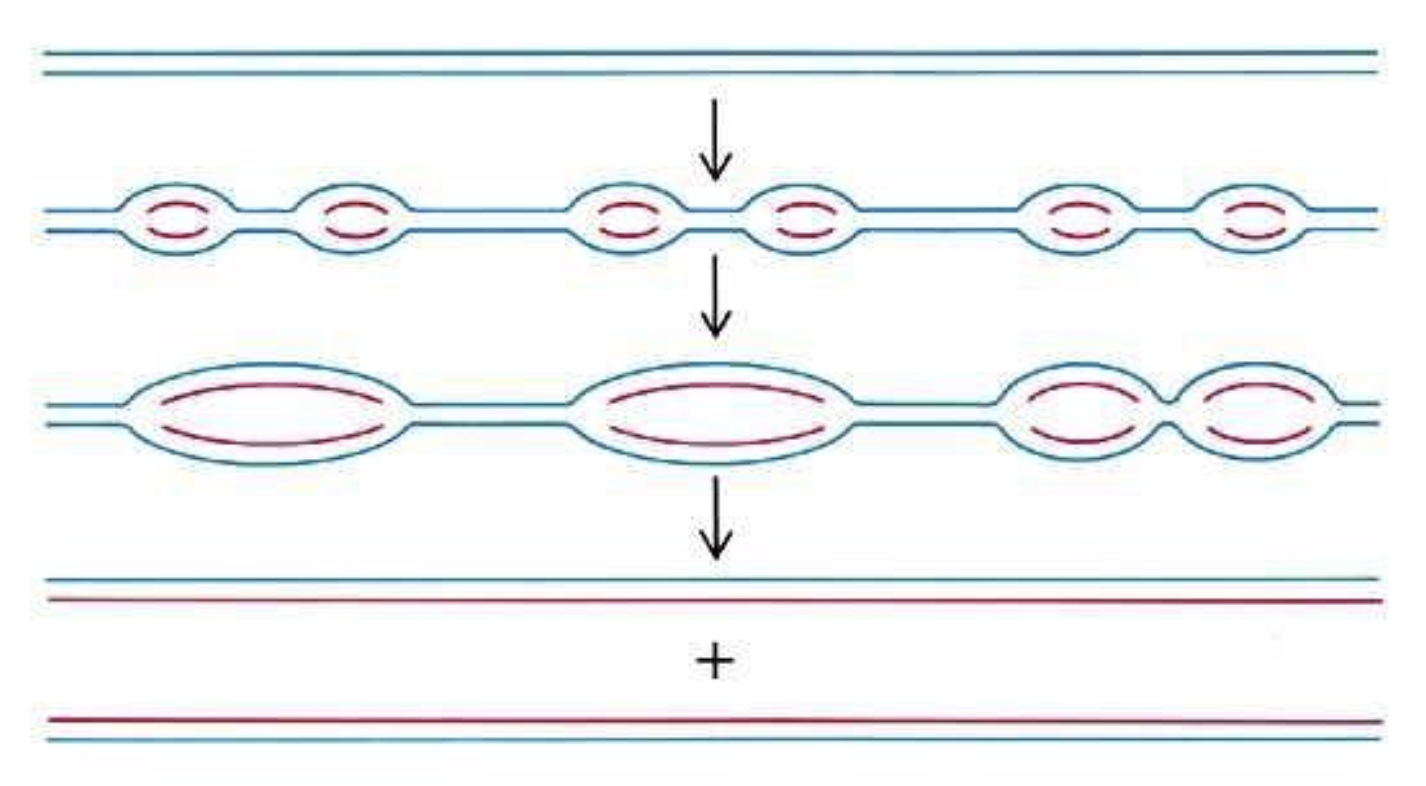
Which phase does DNA get replicated?
S phase
What happens if DNA replication is incomplete?
leads to chromosome breaks during cell division
What happens when over-replication occurs?
leads to extra copies of regions of chromosomes
Does helicase need to be loaded in eukaryotic replication?
no, they’re already loaded and waiting for initiation to start
What is the biggest difference between eukaryotes and bacteria?
eukaryotes have linear chromosomes; bacteria have circular chromosomes
What does eukaryotes having linear chromosomes lead to?
the end replication problem
What causes the end replication problem?
eukaryotes have linear chromosomes
Does the end replication problem occur in bacteria?
no, because bacteria have circular chromosomes, and this problem only occurs due to eukaryotes having linear chromosomes
What is the end replication problem?
the synthesis of the lagging strand results in incompletely replicated DNA due to RNA primer being removed from the last Okazaki fragment but there isn’t a 3’ end to synthesize DNA to replace the removed RNA ∴ if this problem isn’t resolved, as DNA replicates, it gets shorter and shorter
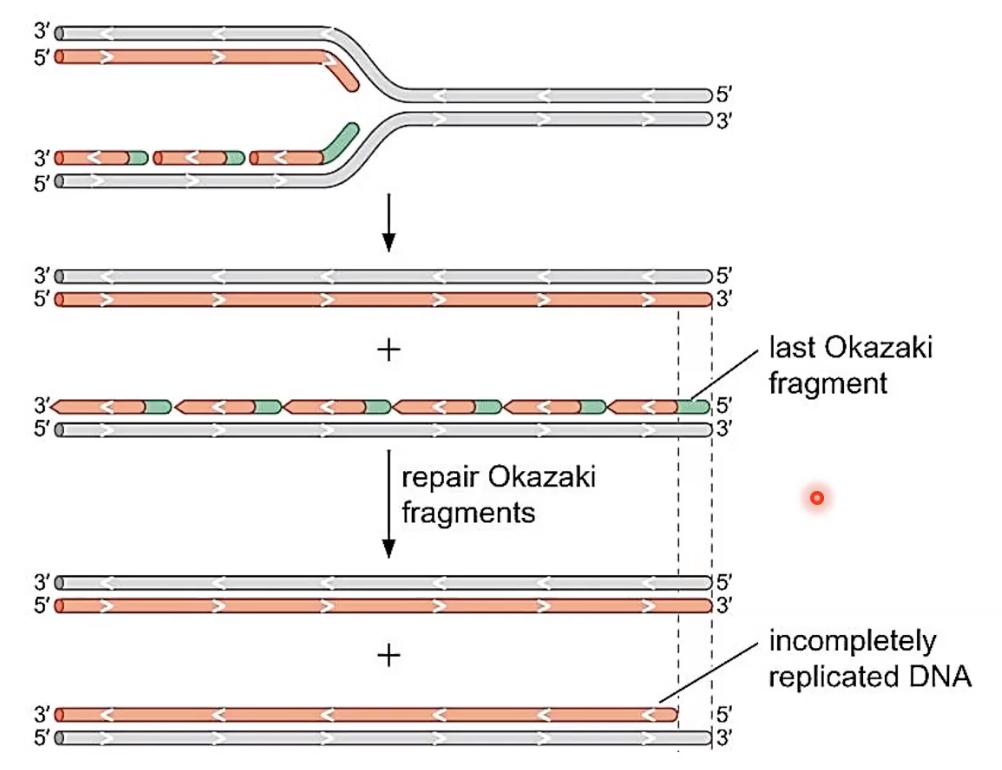
How is the end replication problem compensated for?
telomerase performs “reverse transcription” and makes a DNA strand by copying an RNA strand
What are telomeres?
the ends of eukaryotic chromosomes
What are telomeres made up of?
tandem repeats of TG rich sequences
What do telomeres do?
they protect the ends of linear chromosomes from degradation and other bad things
What is telomerase?
a ribonucleoprotein
a “reverse transcriptase”
What does telomerase do?
creates telomeres
it makes a DNA strand by copying an RNA strand → adds dNTPs to the 3’ end of a primer but its template is RNA, not DNA → compensates for the end replication problem
How is telomerase different from other DNA polymerases?
telomerase uses RNA as template, whereas other DNA polymerases use DNA as template
Do higher eukaryotic cells express telomerase?
yes, but not sufficient enough to repair telomeres
What is senescence?
aging of cells → being unable to repair telomers
Why do cells experience senescence?
they don’t express levels of telomerase sufficient enough to repair telomeres
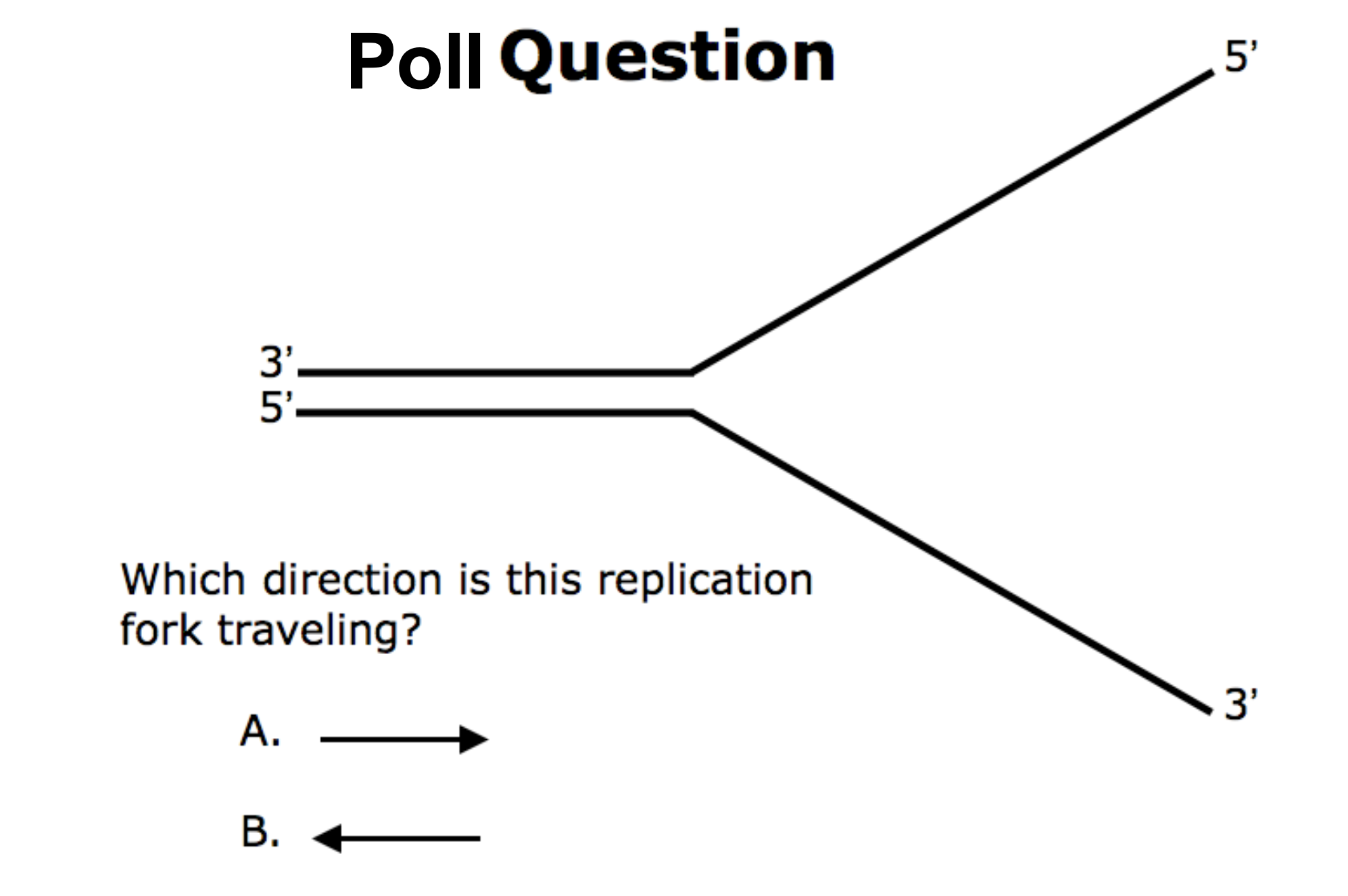
B
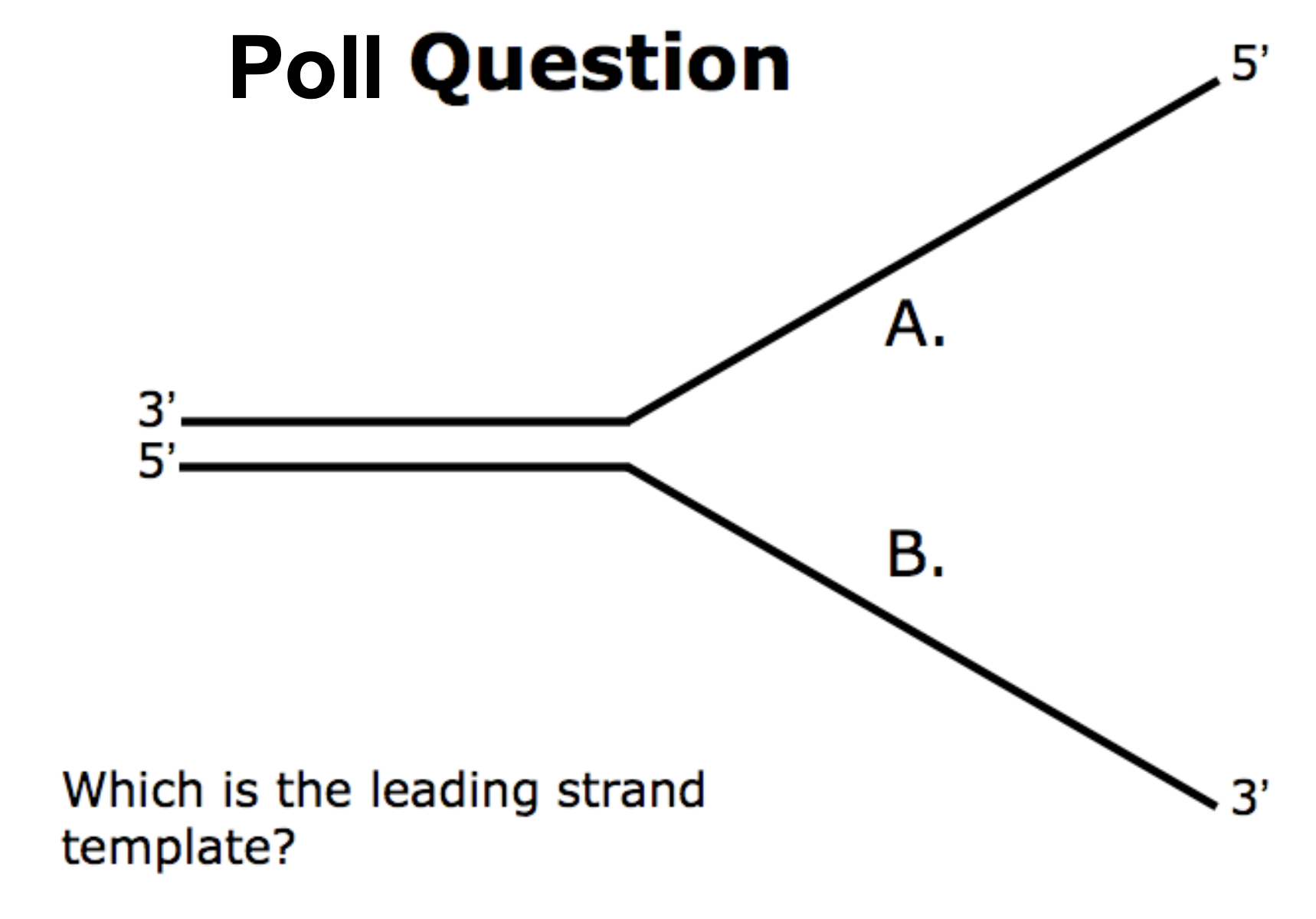
B → leading strands allows synthesis from 5’ to 3’
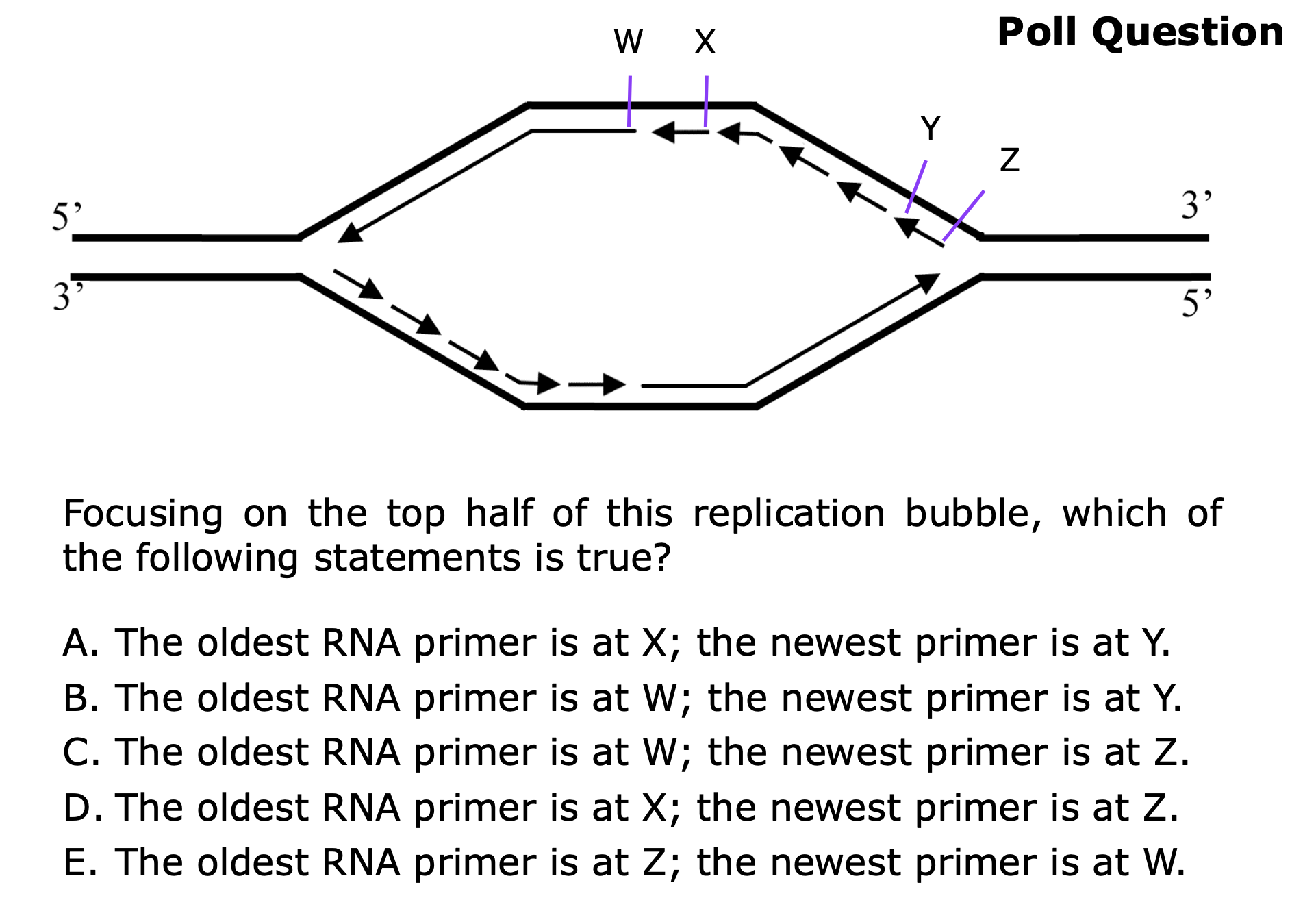
C. The oldest RNA primer is at W; the newest primer is at Z.