Genetic Biology Mc Stay Lectures 1-5
1/123
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
124 Terms
Fundametals of chromosomes
Each human cell comprises of 2 genomes:…
gene encoding regions?
organisations?
Nuclear genome and Mitochondrial genome
Nuclear - only small part encodes genes
-coding elements are spread out - complex, flexible genetics
Mitochondrial genome
-Almost ENTIRELY gene encoding
Highly compact - simple minimalistic
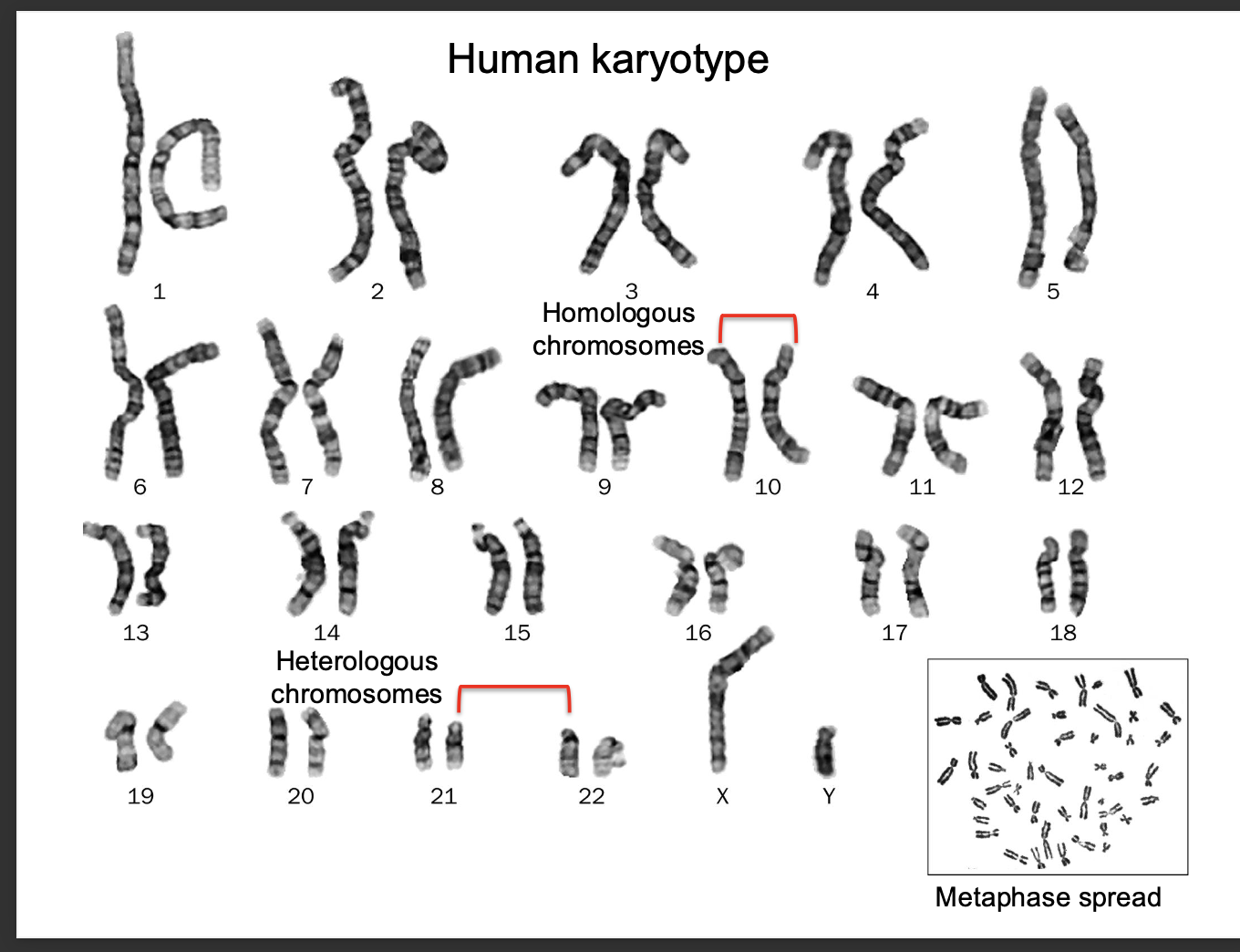
Human karyotype
The complete set of chromosomes in a human cell
shows size,shape structure
1-22 are autosomes
23 - sex chromosomes
-females = 2 x chromosomes
-Males = (x,y)
Karyotypes - used to study genetic abnormalities
such as DOWN SYNDROME - extra copy of chromosome 21 (triosomy)
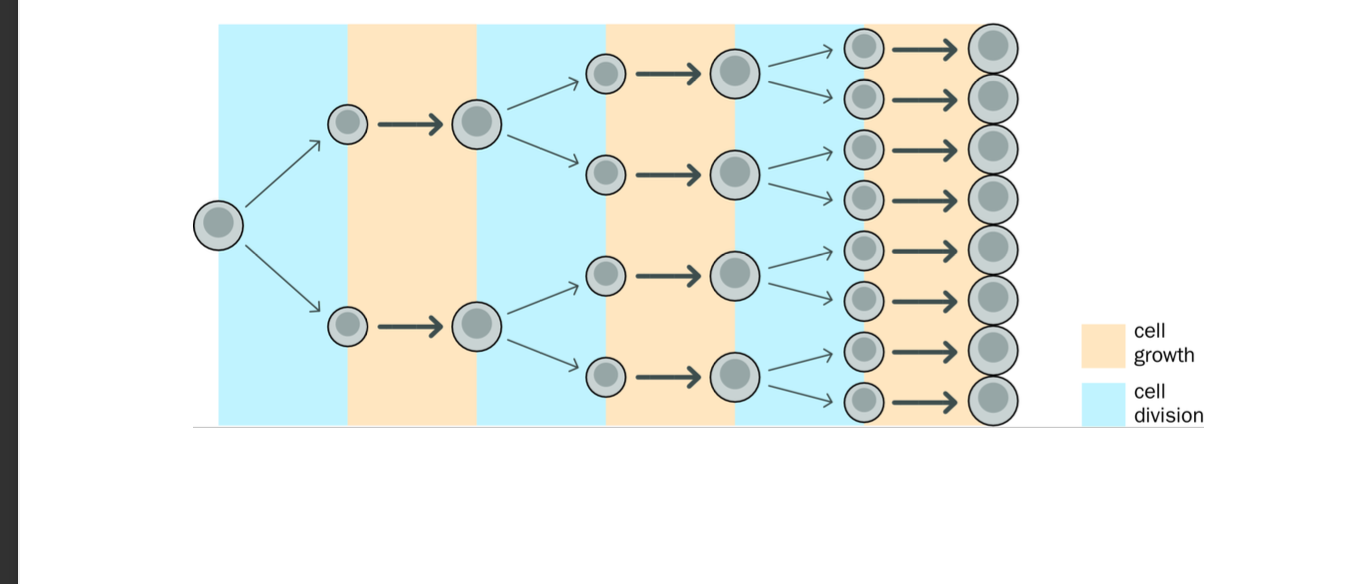
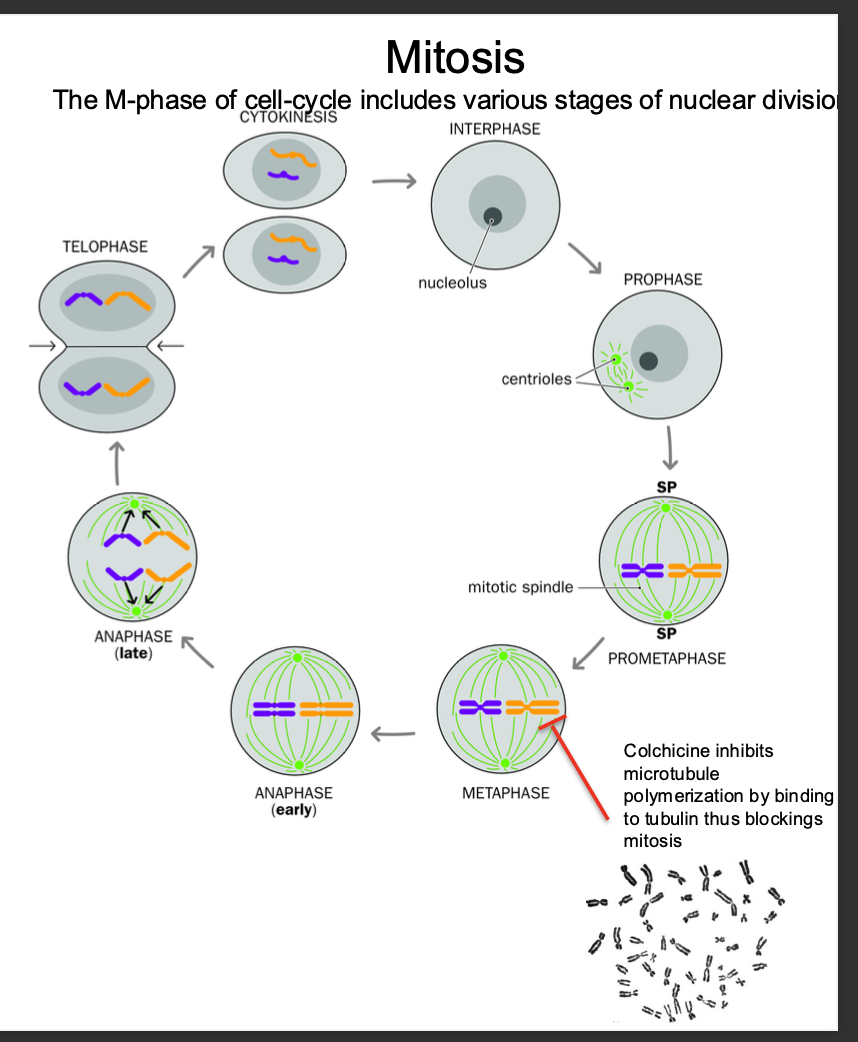
Mitosis
cell division
-Results in 2 genetically identical daughter cells
-each have same number of chromosomes as parent cell
-Mitosis
-Ensures BOTH daughter cells have an IDENTICAL GENOME to that of the parent cell - barring errors
Crucial for growth, development and tissue repair
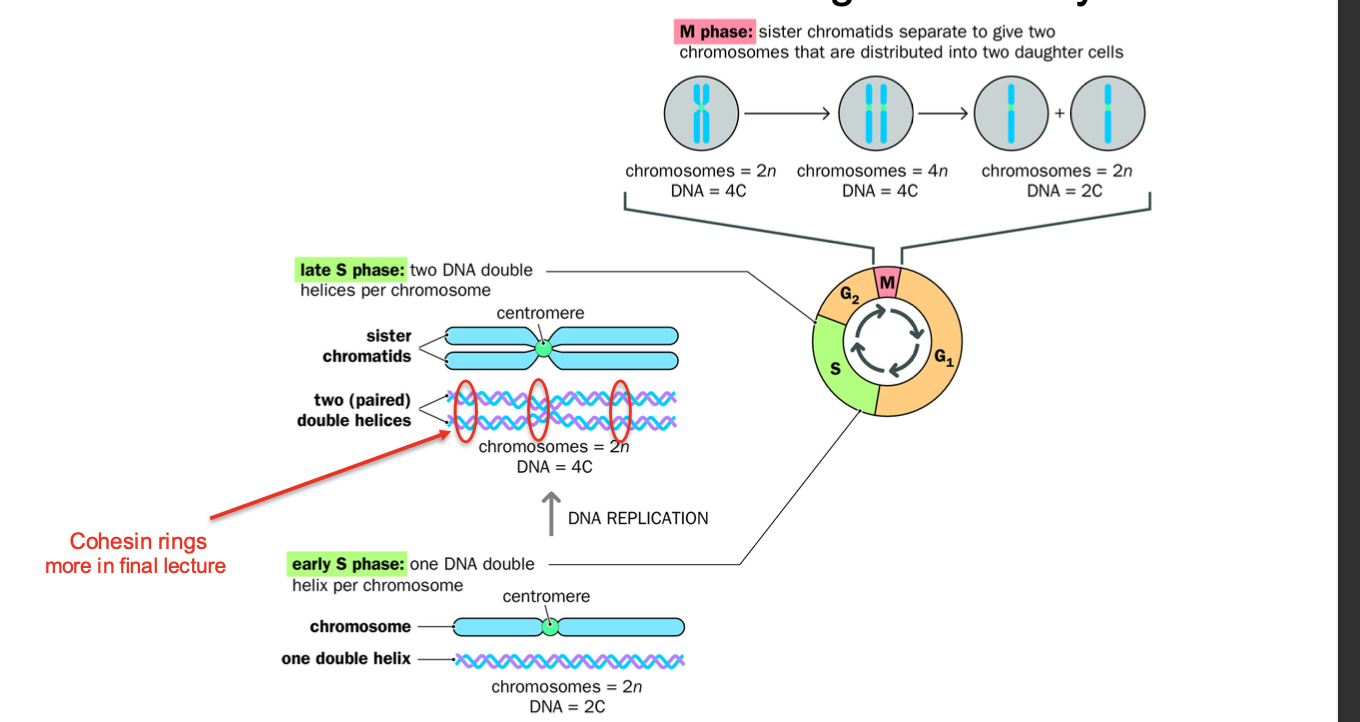
Phases of the cell cycle are….
Explain the phases E
Cell cycle
-Series of events leading to cell DIVISION AND REPLICATION
replication first - makes the sister chromatids - THEN MITOSIS
M phase: Cell division phase - Mitosis and Cytokinesis -CHROMOSOMES ARE CONDENSED
Stages in mitosis: Prophase,prometaphase metaphase,anaphase,telophase,cytokinesis )
Chromosomes condense in preparation for nuclear and cell division
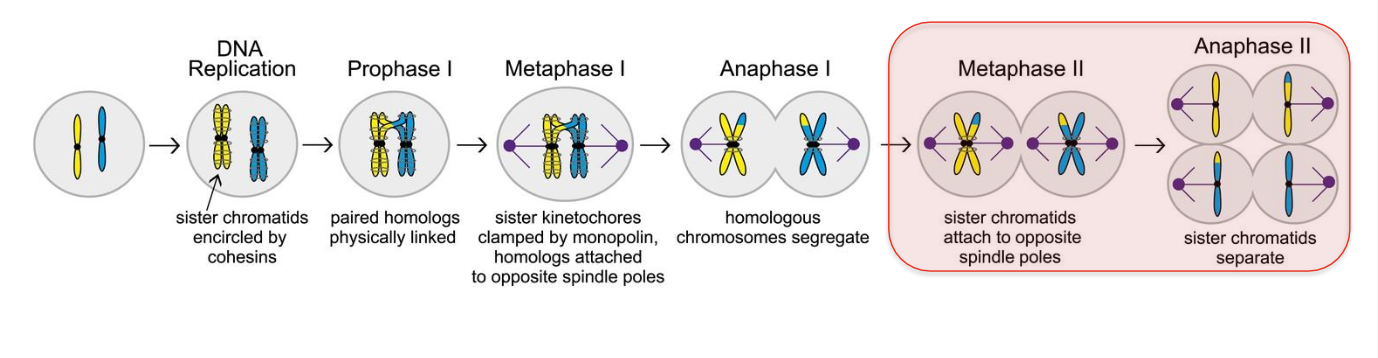
After replication - each chromosome consists of 2 SISTER CHROMATIDS joined by centomere - bound by cohensins
Chromatids aligned along metaphase plate - PULLED APART DURING ANAPHASE- cohesins cleaved
Pulled to opposite poles
Cytokinesis - Cytoplasm divides - forming 2 daughter cells - each with a full set of chromosomes
(SISTER CHROMATIDS SPLIT FORMING 2 DAUGHTER CELLS)
Interphase: Growth + DNA replication - CHROMOSOMES ARE DECONDENSED
G1 - period of cell growth, cell ensures it has the nutrients required to grow
S PHASE:
Dna replication phase - replication of Chromosomes - form - held together by cohesins to be cleaved during anaphase
G2 - further growth and preparation for MITOSIS
Explain the structure of the chromosomes and the behaviour with each phase
Before s phase
Chromsomes contain ONE double helix
-Relaxed + decondensed
After s phase
Sister chromatids - each containing a DNA DUPLEX (double helix)
Each chromatid - contains a single dna double helix
Chromatids joined tightly at the centromere with COHESINS
-allow them to function as a single chromsome
During m phase
-Chromsomes condense
-Align at metaplate
-Cleaved cohesins during anaphase to separate the sister chromatids into individual chromsomes with IDENTICAL genomes
-EQUAL DISTRIBUTION of independent chromosomes after anaphase.
After replication what occurs>
Replication - sister chromatids made
Mitosis now
-Separation of the sister chromatids - held together by cohesin complexes
-Chromosomes condense (shorten + thicken)
Centrioles migrate to opposite poles+ form spindle poles
Prometaphase - NUCLEAR MEMBRANE breaks down - allows for anaphase to occur but also for chromosomes to align along the metaphase plate
Metaphase - residual cohesins removed
-Chromosomes aligned along metaphase plate - cohesins and centromere hold sister chromatid together.
Anaphase - spindles pull sister chromatids apart to opposite poles
Telophase:
reformation of the nuclear membrane around daughter nuclei
-Chromosomes decondense
cytokinesis - separation of the cytoplasm
creates 2 daughter cells
G1 - S PHASE -G2 -M PHASE
ACTIN + MYOSIN CONTRACTILE RING PINCHES CELL
Sexual reproduction
-Genetic mutation was slow prior to this
sex - adaptive feature - common to almost ALL multicellular organisms
-Extremely efficient mechanism for PRODUCING VARIATION
Allowing organisms to ADAPT to changing environments.
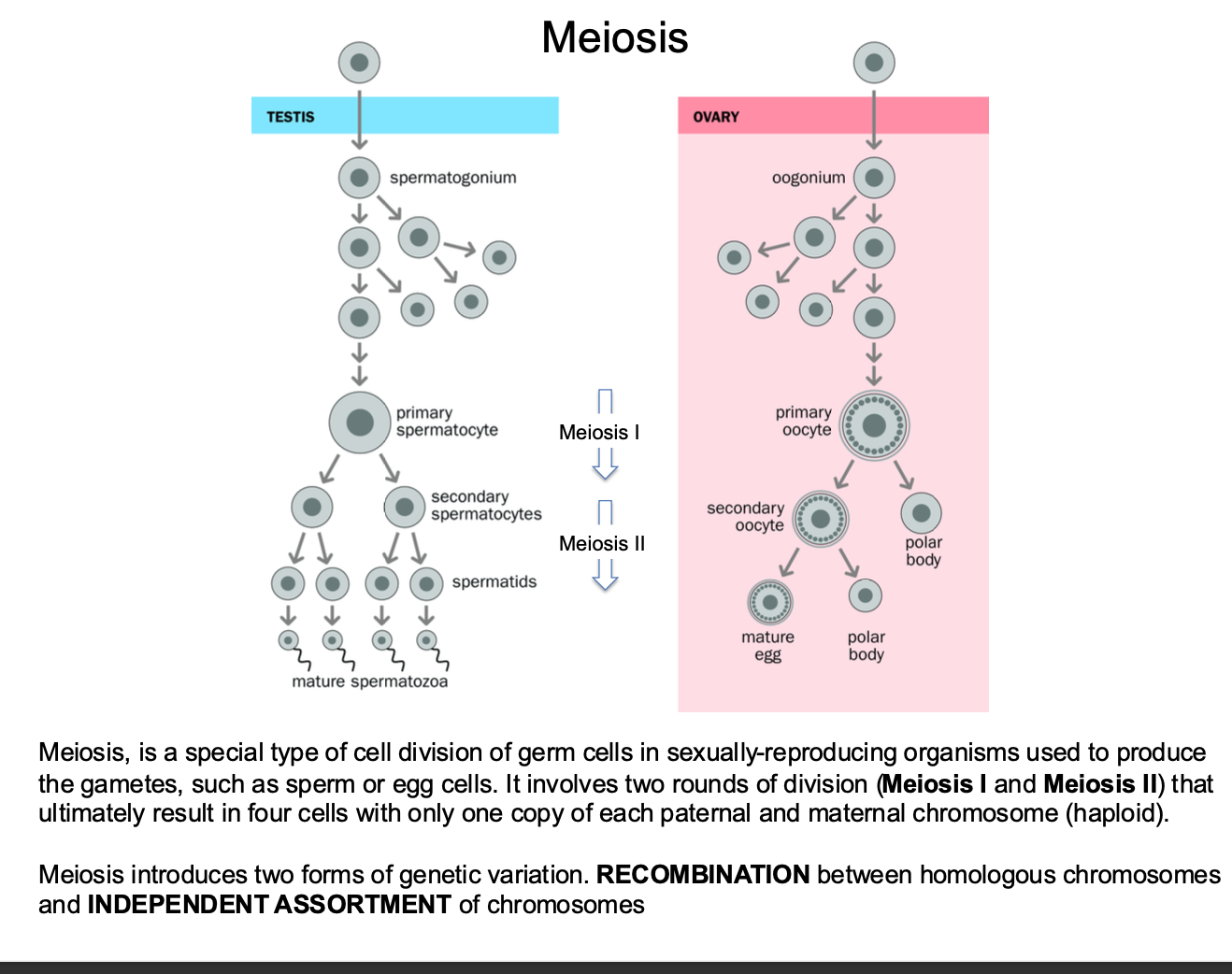
Meiosis is key
-Cell division - iN SEX CELLS
What is meiosis in terms of sexual reproduction:
How many rounds of division
Variation?
Cell division - sexually reproducing organisms use it to produce GAMETES - sperm and egg cells.
2 round of division occur
Meiosis I and Meiosis II
Ultimately result in 4 CELLS with one copy of each paternal and maternal chromosome
2 forms of genetic variation introduced
-Recombination - between homologous chromosomes
-Independent Assortment of chromosomes
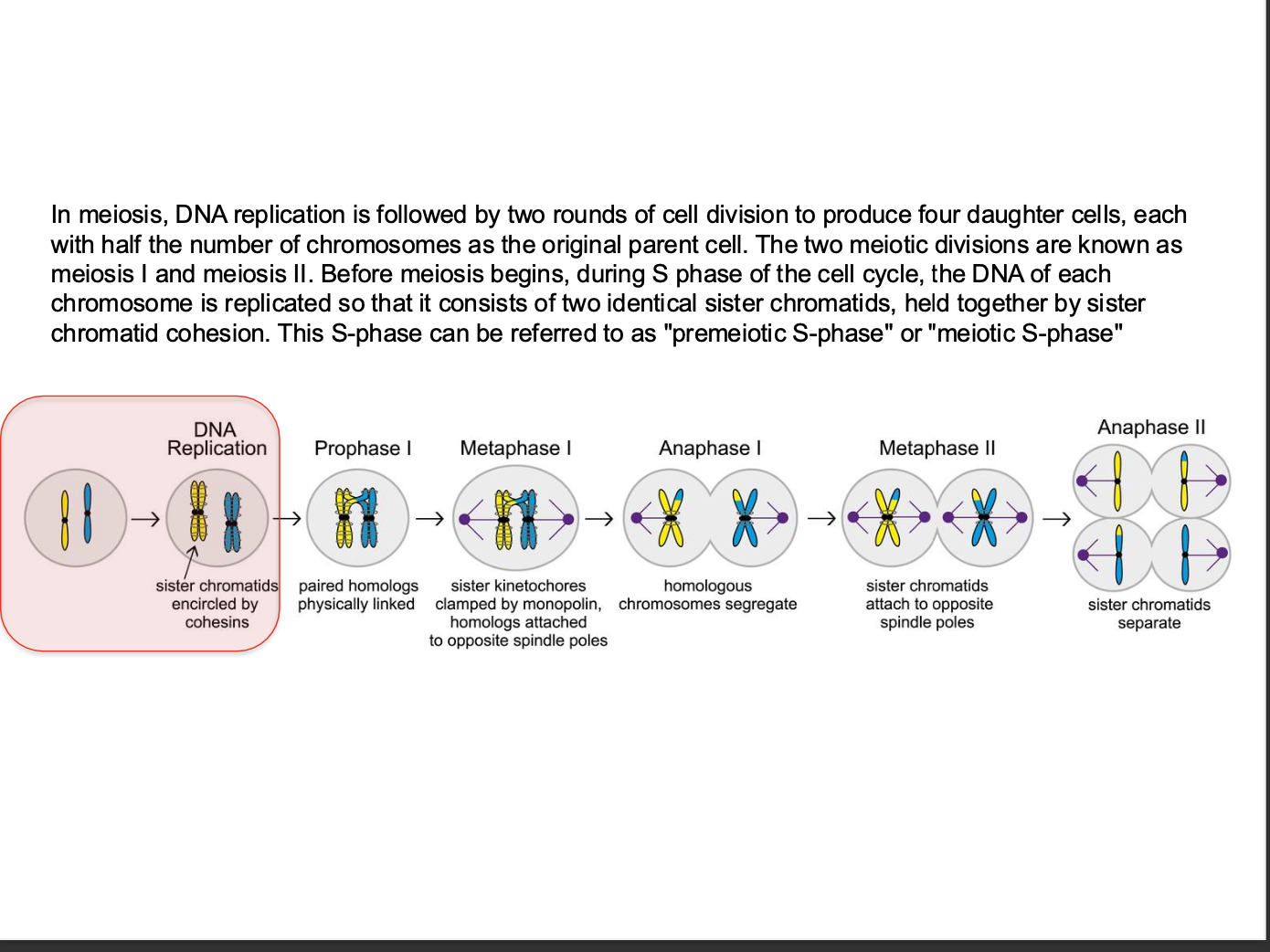
Meosis - how many rounds of cell division
Before or after replication
-how many chromosomes?
What happens before meosis
What is S - phase referred to as?
Meiosis - Specialised reproductive cell division
-During meosis - genetic combinations are randomly created
-Exchanging sequences between maternal and paternal chromosomes BUT ALSO by independent assortment.
AFTER replications - produces GAMETES
2 rounds - 4 DAUGHTER cells - ½ the chromosomes of the parent cell (gametes fuse to get full no of chromosomes)
Before meiosis - replication
2 identical sister chromatids - held together by sister chromatid cohesion
s phase = Premeiotic S - phase
Monopolin homologs - ensure regulated separation of sister chromatids
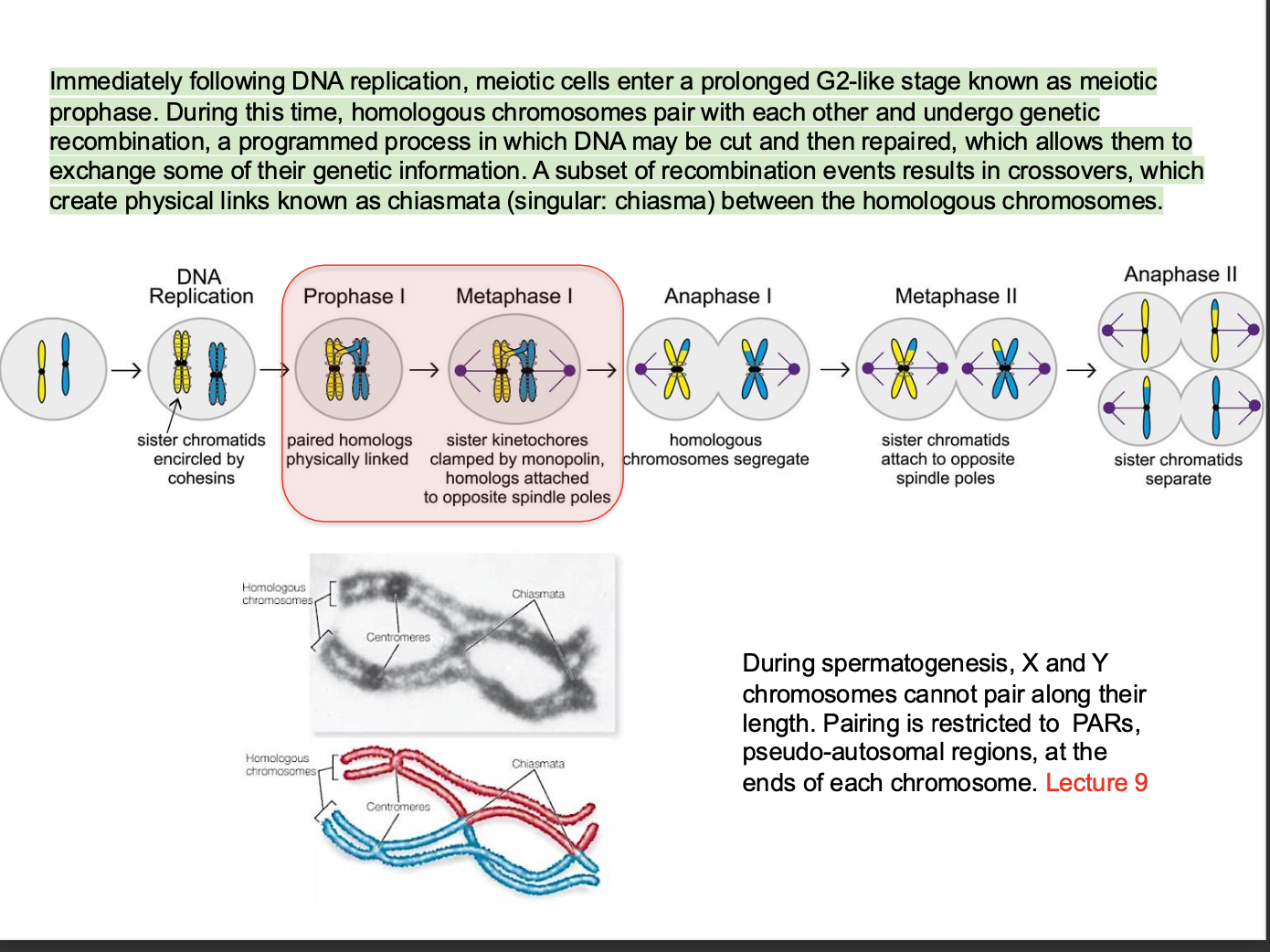
What happens IMMEDIATELY AFTER DNA replication?
over view
Think G1 - S -G2 -M
After s = G2 phase
MEOSIS - Sex cell division - formation of gametes
Meiotic prophase - (prolonged G2 stage)
GENETIC RECOMBINATION
Homologous chromosomes PAIR + UNDERGO genetic recombination
(One chromsomes from the mother and one from the father pair up) - Synapsis
During spermatogenesis X and Y chromosomes CANNOT PAIR ALONG THEIR LENGTH
Pairing only occurs at pseudo-autosomal regions at the end of each chromosome
Recombination process involves
Cutting + Repairing of DNA - allowing exchange of genetic information. - specialised enzymes create double strand breaks in the DNA of the chromatids of homologous chromosomes.
After the breaks - DNA is repaired by exchange of chromatids between homologous chromosomes - Results in the MIXING OF GENETIC MATERIAL between the mother and the father - creating
Recombination:
Part of one of the maternal chromosome is swapped with part of the paternal homologous chromosome
Cross overs:
Segments of chromatids from homologous chromosomes are exchanged
Chromatids exchange genetic material through chiasmata - Physical links in homologous chromosomes where genetic material is exchanged
RECAP: GENETIC diversity - meiosis - genetic recombination
Meiosis - Specialised reproductive cell division
-During meosis - genetic combinations are randomly created
-Exchanging sequences between maternal and paternal chromosomes BUT ALSO by independent assortment.
Prophase I
Meosis - sex cell division
STAGE 1
How is recombination initiated?
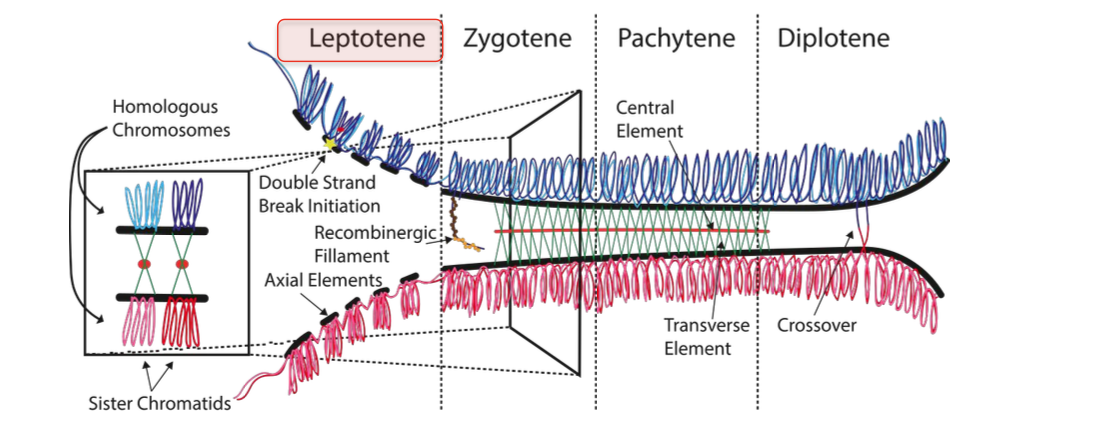
LEPTOCENE
-Formation of synaptomenal complex - condensing of chromsomes
-Double strand break initiation by SPO11
Longest phase of meiosis
Chromatid exchange
-Maternal and paternal chromsomes pair, synapse + exchange genetic information - BY GENETIC RECOMBINATION (break + repair)
-Form at least ONE crossover per chromosome
Stage 1
-Leptotene stage
Chromosomes condense - become visible under a microscope
-Sister chromatids held together by cohesin
Chromosome individualisation occurs - Individual chromosomes become distinct visible threads
Chromosomes arranged as array of loops - loops extend from the AXIAL ELEMENT
Condensing of chromosomes - forms synaptonemal complexes
-helps tight pairing of chromosomes
Lateral elements of the synaptonemal complex form along each chromosome - forming the axial element - from this loops of the chromosomes extend - each containing a portion of chromosomal DNA
Synaptomenal complex - crucial for correct alignment of chromosomes for recombination
Chromosome organisation
Allows chromosomes to find and pair with their homologous partner in ZYGOTENE
LEPTOTENE - aids recobmination - exchange of genetic information leading to genetic diversity
DOUBLE STRAND BREAK INITIATION (needed for genetic recombination to occur)
Enzyme SPO11 creating programmed double strand breaks
Step 2 in prophase meiosis
Zygotene
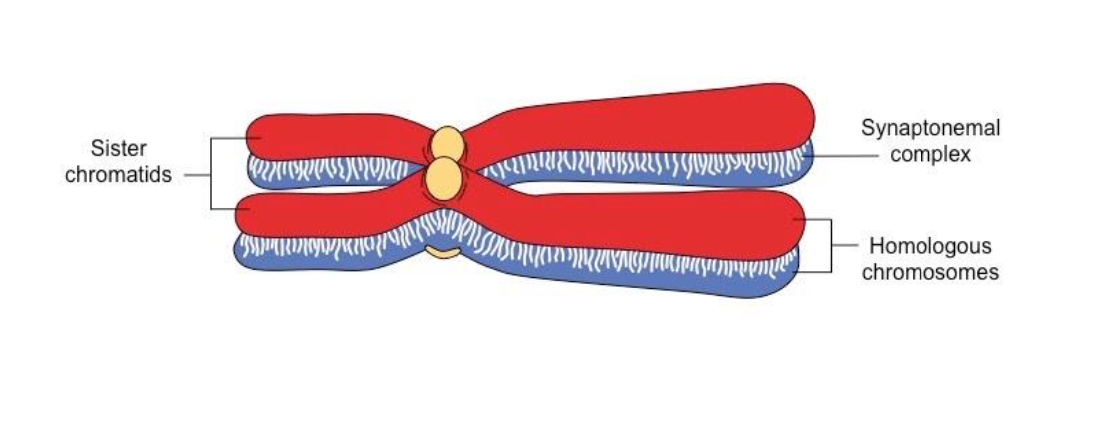
-CHROMOSOME PAIRING - BIVALENT FORMATION
Synaptomenal complex is formed
-Homologous chromsomes are much more closely and stably paired - Due to the formation of the synaptomenal complex
-2 chromosomes paired together - form bivalent structure - stabilised by s.c.
prepared for genetic recombination
Recombination is already beginning with pairing stabilised by s.c
Prophase step 3
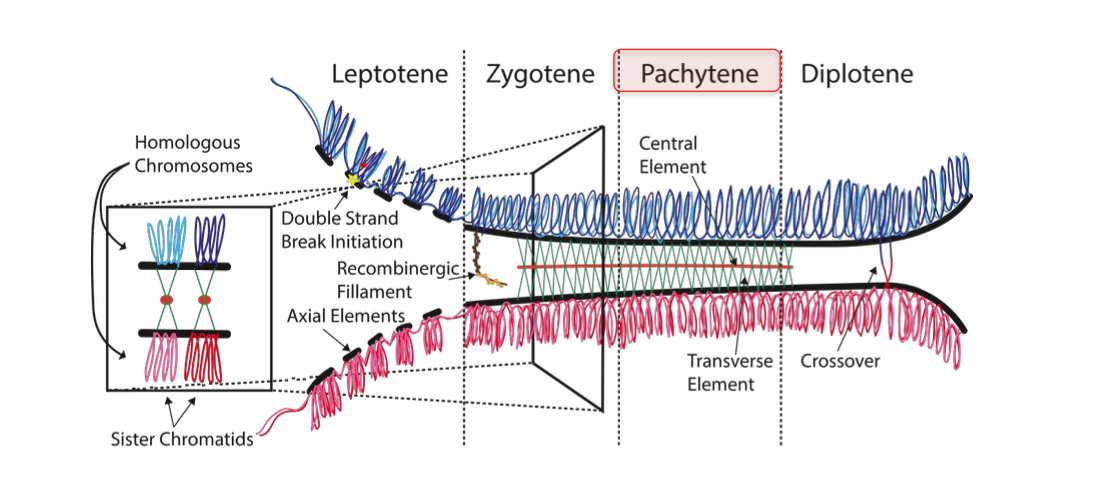
Pachytene
(patching up the breaks)
completion of crossing over + genetic recombination
Repair of double strand breaks
-Homologous recombination - including chromosomal cross over
-Completion - by REPAIR of DOUBLE STRANDED BREAKS created in leptotene
Most repaired WITHOUT FORMING CROSS OVERS - At least one cross over per chromosome
Subset of breaks cause cross over -
-Cross over / exchange of genetic material between non sister chromosomes (non homologous)
Causes genetic diversity in offspring
prophase step 4
Diplotene
SLIGHT SEPARATION - still connected in some parts (chiasmata) - chromosomes become more DI
-Synaptomenal complex disassembles - homologous chromosomes SEPARATE A LITTLE
Homologous chromosomes of each bivalent
-Remain tightly bound to chiasmata - the regions where crossing - over occured
Chiasmata - ensure correct chromosome segregation - without them - nondisjunction could occur ( improper chromosome segregation)
Chiasmata remain on the chromosomes until they are SEVERED at the transition to anaphase I - to allow the homologous chromosomes to separate + move to opposite poles of the cell
Prophase steps
L,Z,P,D. - THINK lspd but z
Anaphase of meiosis
2 PARTS
1 AND 2
ANAPHASE - MEIOSIS ONE
HOMOLOGOUS CHROMOSOME SEPARATION
-homologous chromosomes - segregate away from each other
-2 HAPLOID CELLS ½ number of chromosomes of the parent cell
gre
ANAPHASE - MEIOSIS TWO
Sister chromatids segregate - COHESION BETWEEN SISTER CHROMATIDS IS RELEASED
Segregate from each other
4 haploid cells
Meiosis 2 differs to mitosis because
chromsomes have mix of genetic information due to recombination in prophase
MEOSIS GENETIC VARIATION
-Genetic recombination between homologous chromosomes
(as discussed)
-prophase - leptocene, zygotene, pacytene, diplotene
Break and repair - cross over and recombination
segregation - meiosis 1 and 2 = homologous chromosomes separate 2 = cohesion removed sister chromatids separate 4 cells ½ number of chromosomes of parent
REMEMBER GAMETES HAVE ½ NUMBER OF CHROMOSOMES
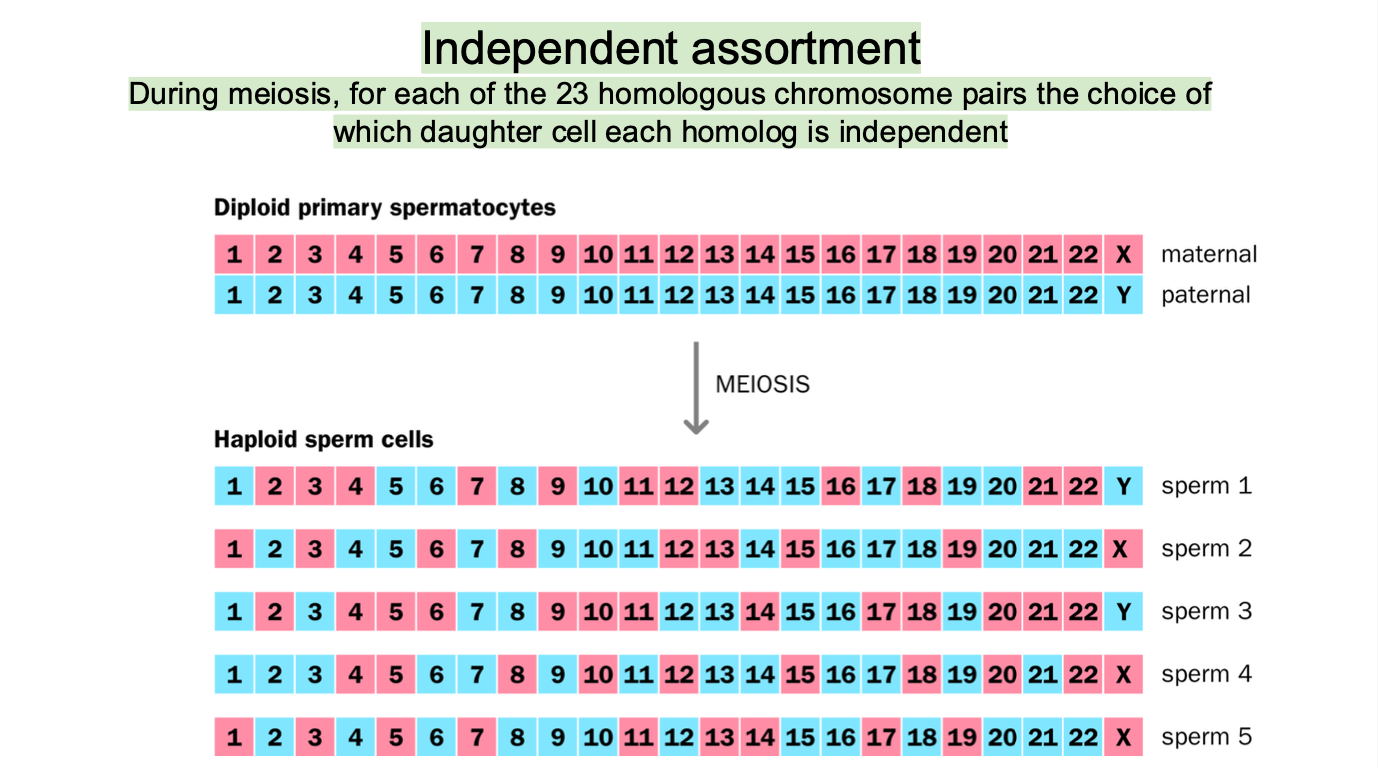
OTHER MODE OF VARIATION - Independent assortment
independent assortment
The 23 chromosomes are aligned and separated in such a way that the combination of MATERNAL AND PATERNAL chromosomes in the resulting gametes is random
SPERM AND EGG CELLS HAVE A MIX OF CHROMOSOMES
Homologous chromosomes are RANDOMLY DISTRIBUTED to different daughter cells during anaphase 1 of meiosis
(ONE FROM MOM AND ONE FROM DAD)
23 CHROMOSOMES
2²³ COMBINATIONS possible
-High genetic diversity
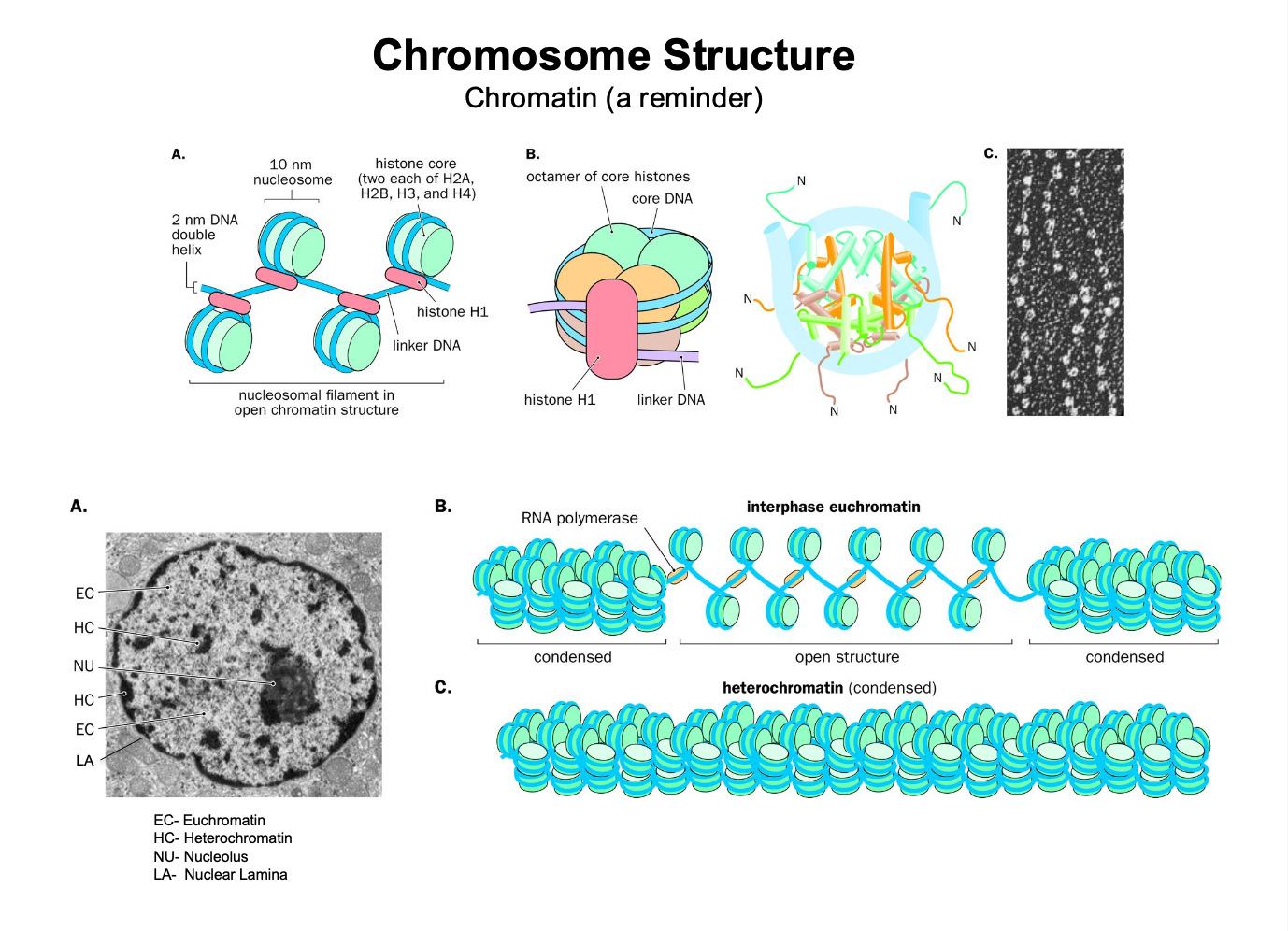
Structure of chromosome recap
The dna inside the chromosome is compacted into a chromatin structure
dna in the chromatin structure is wrapped around octamer of core histones
-forms a nucleosome
Heterochromatin: - dna is tightly bound to the histone - hard access - inactive transcription
Euchromatin - more relaxed dna - easier to access - active transcription
Interphase - chromosomes are less condensed - NOT tightly condensed
Metaphase - chromosomes are TIGHTLY condensed
mitosis and meiosis - condensing of chromosomes - allows for accurate division - individualisation
autosomes - non sex chromosomes 22 of them
sex chromosomes 1 pair (X,X) OR (X,Y) female / male
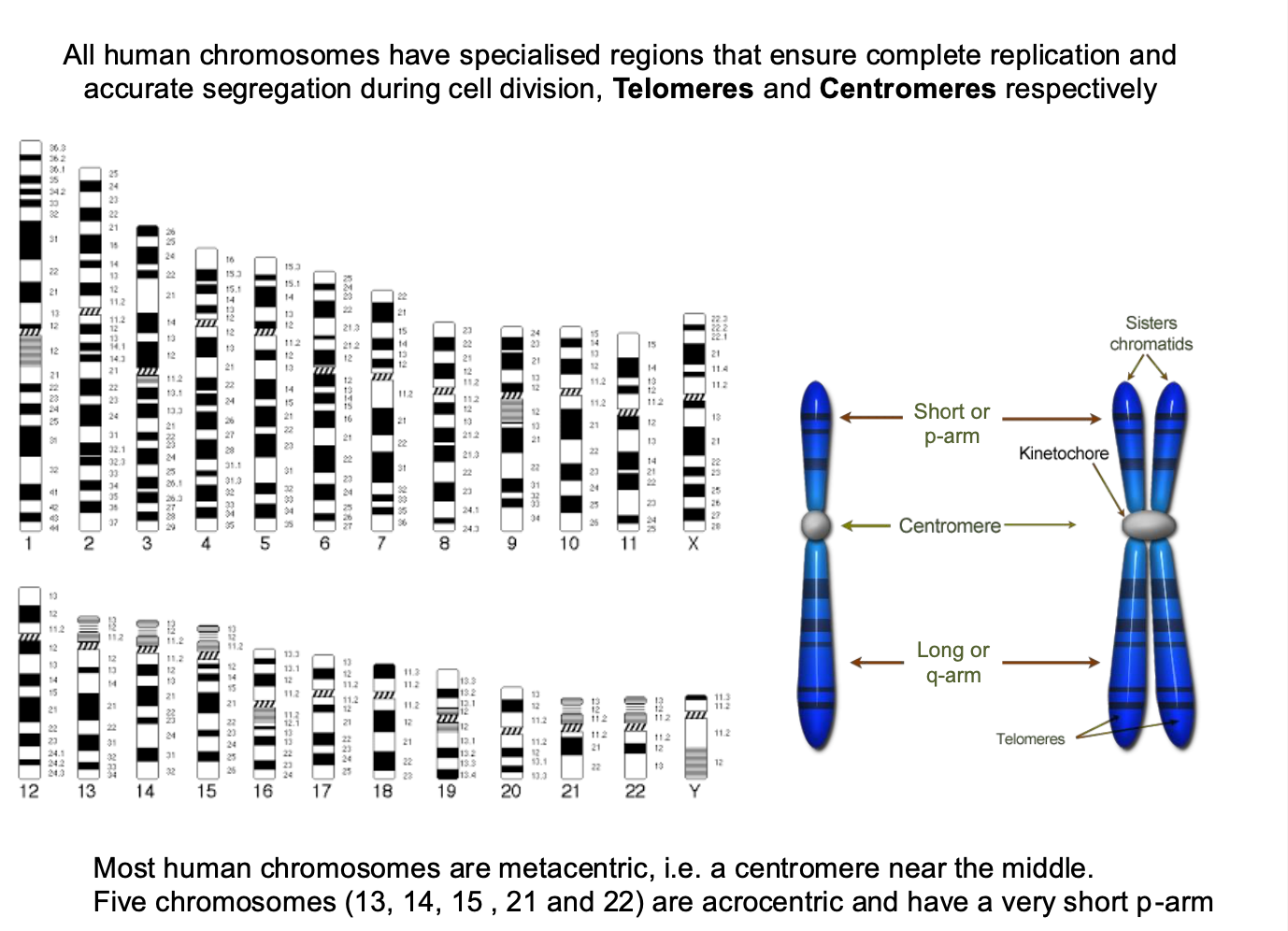
Specialised regions in human chromosomes - what are the functions?
What are most humans
Specialised regions
-Ensure complete replication + accurate segregation during cell division
-centromeres and telomeres
-Most humans are metacentric - centromere near the middle
-5 chromosomes = acrocentric - short p-arm - contains little / no genetic information
submetacentric - slightly off center
Play a role in the genetic function of the chromosome
Down syndrome = trisomy of chromosome 21
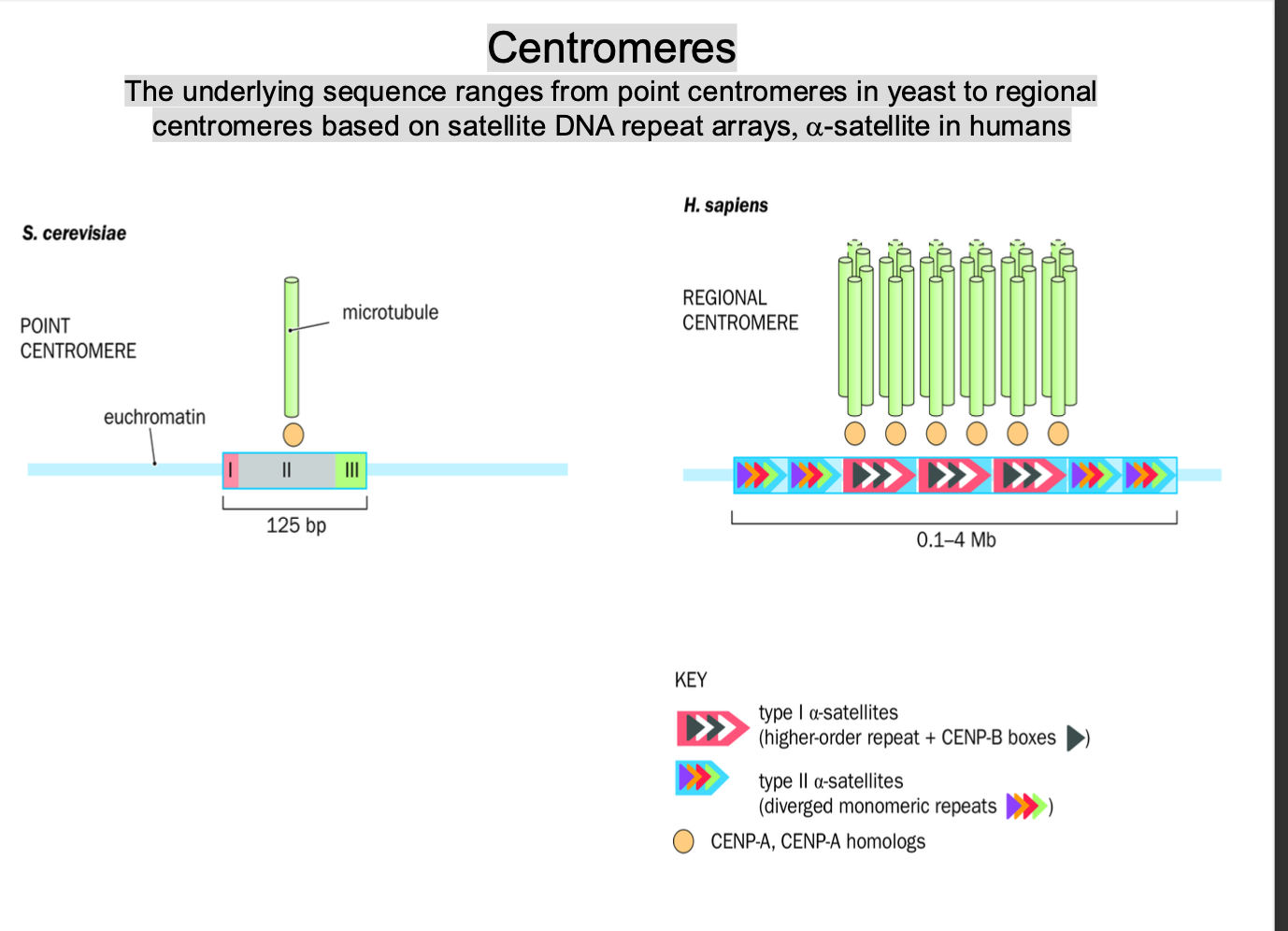
Centromeres
function?
-TYPES / ranges of centromeres
POINT / REGIONAL
SMALL + DEFINED / LARGE + REPETITIVE - RELY ON KINETOCHORES
centromere - region of DNA on the chromosomes - acts as POINT of attachment for the kinetochore
-Crucial during cell division
ensure correct segregation of chromosomes into daughter cells
ranges point - regional -
Point centromeres:
Yeast - least complex
-very short, small, well defined dna
-Specific - dont involve large repetitive DNA
Centromere sequence - essential for attachment of spindle to microtubules + correct chromosome segregation
Regional centromeres:
Higher eukaryotes
-More complex
Large + repetitive regions of sequences of DNA
Long arrays of repetitive dna sequences - satellite dna repeat arrays
Satellite dna
-Non coding
-essential for centromere function + proper chromosome segregation
Alpha satellite dna
-Long tandem repeats
-Necessary for centromere formation
repetitive nature stabilise the centromere + enables the correct attachment of kinetochore proteins involved in chromosome movement
Evolution and Centromere Identity:
In higher eukaryotes, centromeres are not defined by a single, simple DNA sequence but rather by specific chromatin modifications and the presence of kinetochore proteins that allow for correct chromosome segregation.
Centromere identity is not solely determined by the DNA sequence but is also influenced by epigenetic factors, including modifications like histone variants and DNA methylation. This is why centromeres can be shifted or redefined in certain species or conditions.
Yeast (e.g., Saccharomyces cerevisiae): Yeast centromeres are composed of a short, defined sequence (point centromeres) that is crucial for accurate chromosome segregation.
Humans and other mammals: Centromeres consist of long arrays of repetitive DNA sequences, such as α-satellite DNA, and rely heavily on kinetochore proteins for function.
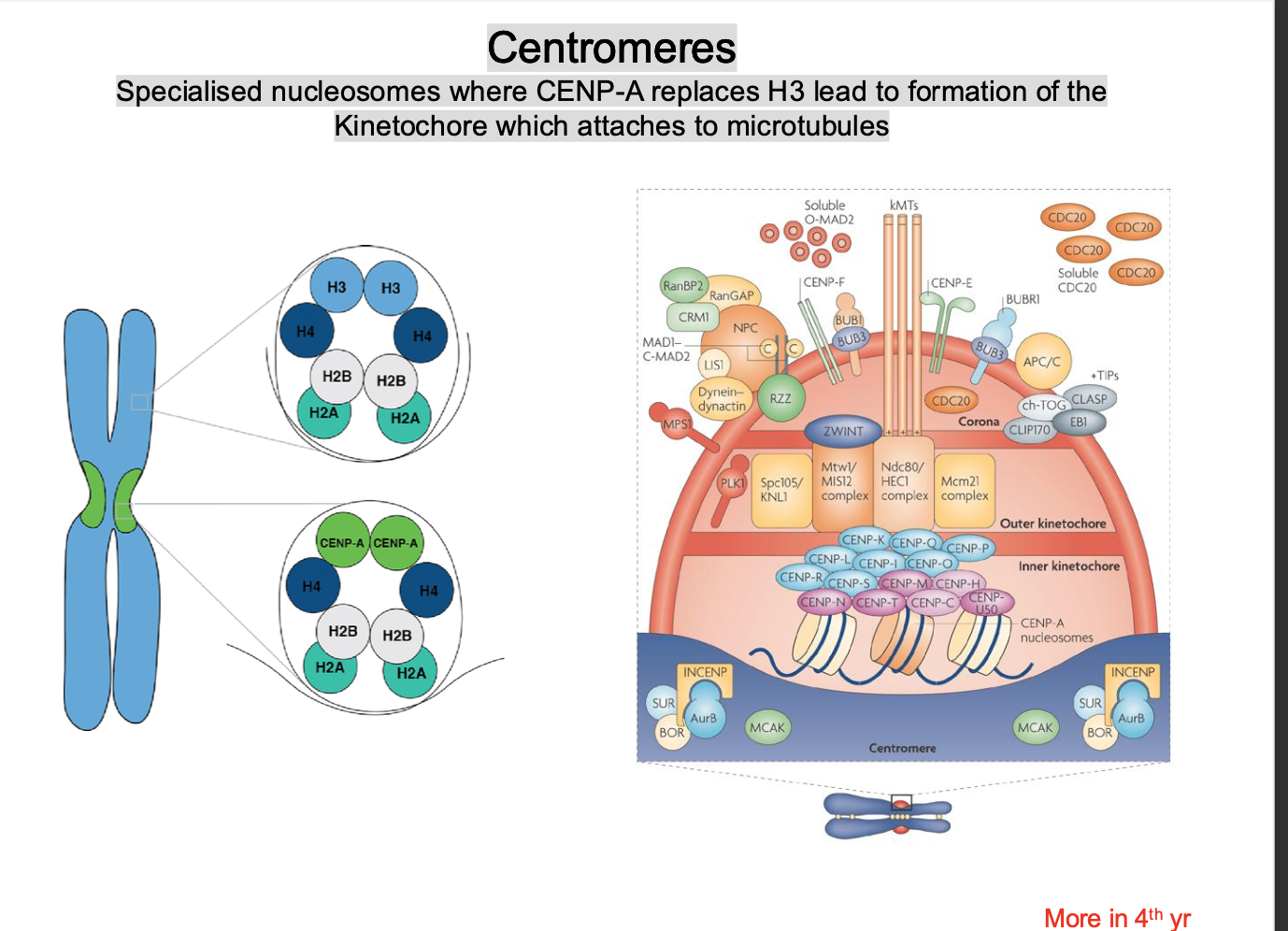
centromeres - Formation of the kinetochore
a histone protein H3 is replaced by a VARIANT HISTONE CENP-A
-stabilises the structure
-Forms on the centromere
-Recruits kinetochore proteins to form the kinetochore
CENP-A acts as a SCAFFOLD for the formation of the kinetochore protein
Telomeres
function
How is this carried out?
-Protect the ENDS of linear chromosomes
-Cap the chromosomes - PREVENT INTERNAL DNA being degraded by nucleases
-solve the end replication problem
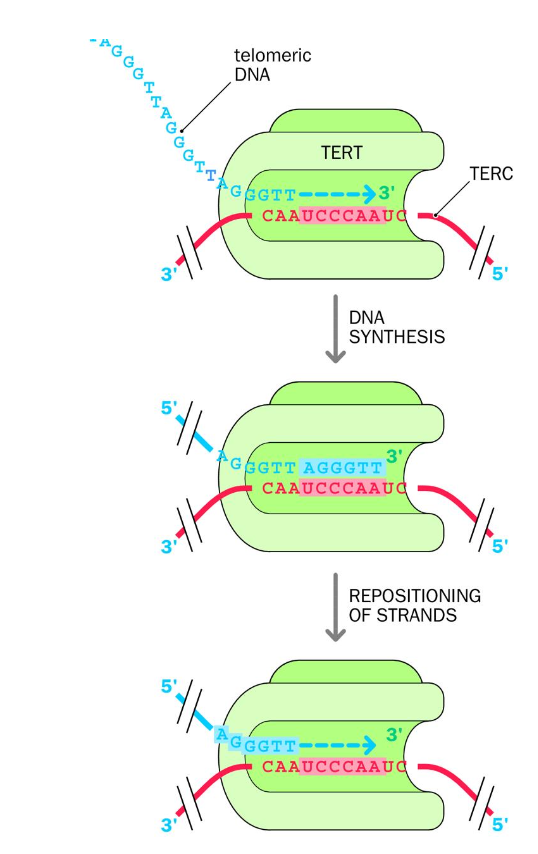
Maintained and extended by telomerase enzyme
Telomeres can shorten with cell division - without telomerase this would cause cell death.- counteracts shortening
telomerase - uses reverse transcriptase + noncoding RNA template to make new TELOMERE dna repeates
-TERT - telomerase reverse transcriptase is an RNA-dependent DNA polymerase - synthesis of DNA from an RNA template
uses RNA template provided by its other subunit TERC - telomerase RNA component.
(terc) The rna template contains a sequence that is complementary to the telomere repeat sequence in humans - allowing tert to use terc to add more dna repeats to the telomere
complementary sequence = hexanucleotide sequence in the rna template
TERT USES TERC to add telomere repeat sequences to chromosome ends
Telomeres
Shelterin complex
telomere = the protective cap on the ends of chromosomes - consists of the shelterin complex - prevents internal dna from being degraded by nucleases
Specialised set of proteins - protects chromosomes ends- prevents them from being interpreted as dna double strand breaks
(genomic instability would ensue)
Telomeres:
TRF1 AND TRF2 - proteins that bind to double-stranded telomeric repeats - responsbile for
TRF2 - lengthening the telomere
TRF1 - protection + forming the protective t- loop that prevents recognition as a double strand break
POT1 - interacts with the single-strand 3’ overhang of the telomere dna - TTAGGG
Binding proteins
TIN2 AND TPP1 -connect POT1 to trf1 + trf2 - help complete the shelterin complex
Rap 1 - binds to trf2 - contributing to stability + protection of the telomere
prevents inappropriate activation of DNA damage response
t-loop creates lariat like structure - physically protecting the chromosome ends
-t -loop shields chromosome ends -prevents them from being recognised as a double strand break
Maintain genomic stability, prevent aging + cancer
importance of centromere and telomeres
-Faithfully transmit DNA from mother to daughter cells
centromere - correct segregation at cell division
telomere - cap the chromosomes - prevents the internal dna from being degraded by nucleases
-prevent inappropriate dna damage response
Lecture 2
Patterns of Inheritance and how to study and identify chromosome
Definitions
Locus
Alleles
Genotype
Phenotypes
Locus - the unique chromosomal location - position of the gene on the chromosome
-defines the position of an individual gene or DNA sequence
ABO blood locus group (example)
Alleles - alternative form of the same gene - a,b,o
genotype - List of the alleles present at one / several loci
phenotype - observable traits / character(istics) of an organisms
-simple inspection to sophisticated lab investigations (range)
more terms:
zygote
homozygous
heterozygous
autosomal / x linked
dominant
recessive
Zygote - union of the sperm cell and the egg cell
Homozygous - if both alleles at the locus are the same
heterozygous - both alleles at the locus are different
Autosomal / x linked character
-depending on the location of the gene on the chromosome
-few y linked characters
autosomal character - on the non sex genes
-not sex-dependent
-can be dominant / recessive
x-linked (not many y linked)
-females can be carriers if they have 1 x chromosome mutation
-More chance for mutation to occur as they have 2 X chromosomes
Males are (X,Y)
-males are most affected - have 1 chromosome (only 1 no back up like females)
Dominant - if heterozygous and CHARACTER OBSERVED
Recessive - if heterozygous and character NOT OBSERVED
Gene
-A functional unit of dna
-a determinant / co- determinant of a character that is inherited
gene encodes one protein - have sequences that encode the protein included in the dna
HUMAN GENES - UPPERCASE ITALICIZED NAMES
Monogenic vs multifactorial inheritance
(genotype = list of alleles at locus)
Monogenic
-Genetic characters whose presence / absence depends on the GENOTYPE AT A SINGLE LOCUS
-Mendelian characters - pedigree patterns
Non-mendelian / MULTIFACTORIAL
-GENOTYPE AT MULTIPLE LOCI
-Genetic characters are governed by genes at MORE THAN ONE locus
-governed by a small number of loci = oligogenic
-governed by lots of loci, each with small effects = polygenic
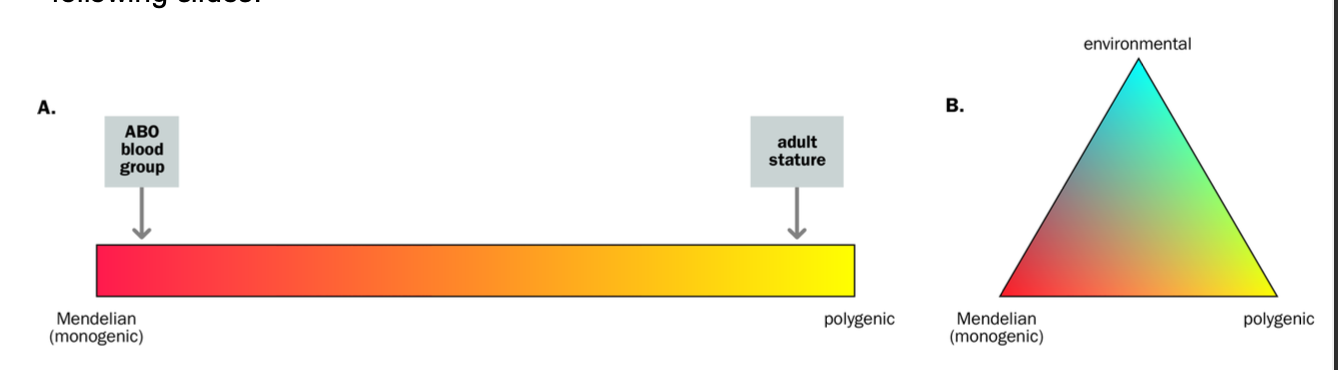
Mendelian characters are usually described as…
-Described as dichotomous - you have it or you don’t e.g. extra finger
Cystic Fibrosis
HUMAN TRAITS
-Non - mendelian - not dichotomous
Most a CONTINUOUS / QUANTITATIVE CHARACTERS e.g. height / weight
-We all have them, just to different degrees
Continuous traits - QTLs - quantitative trait locis
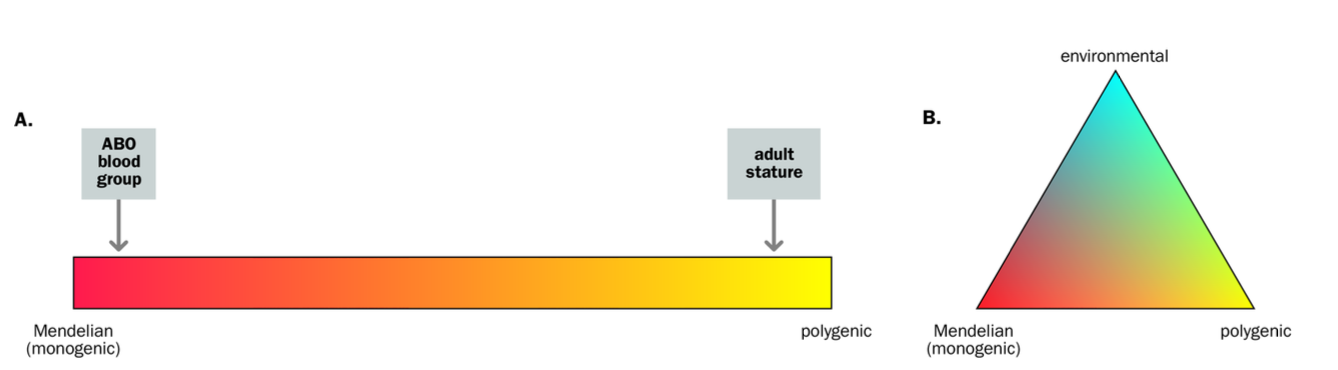
mendelian pedigree patterns
square - man
circle = woman
dot = carrier
blue = affected
single line = mating
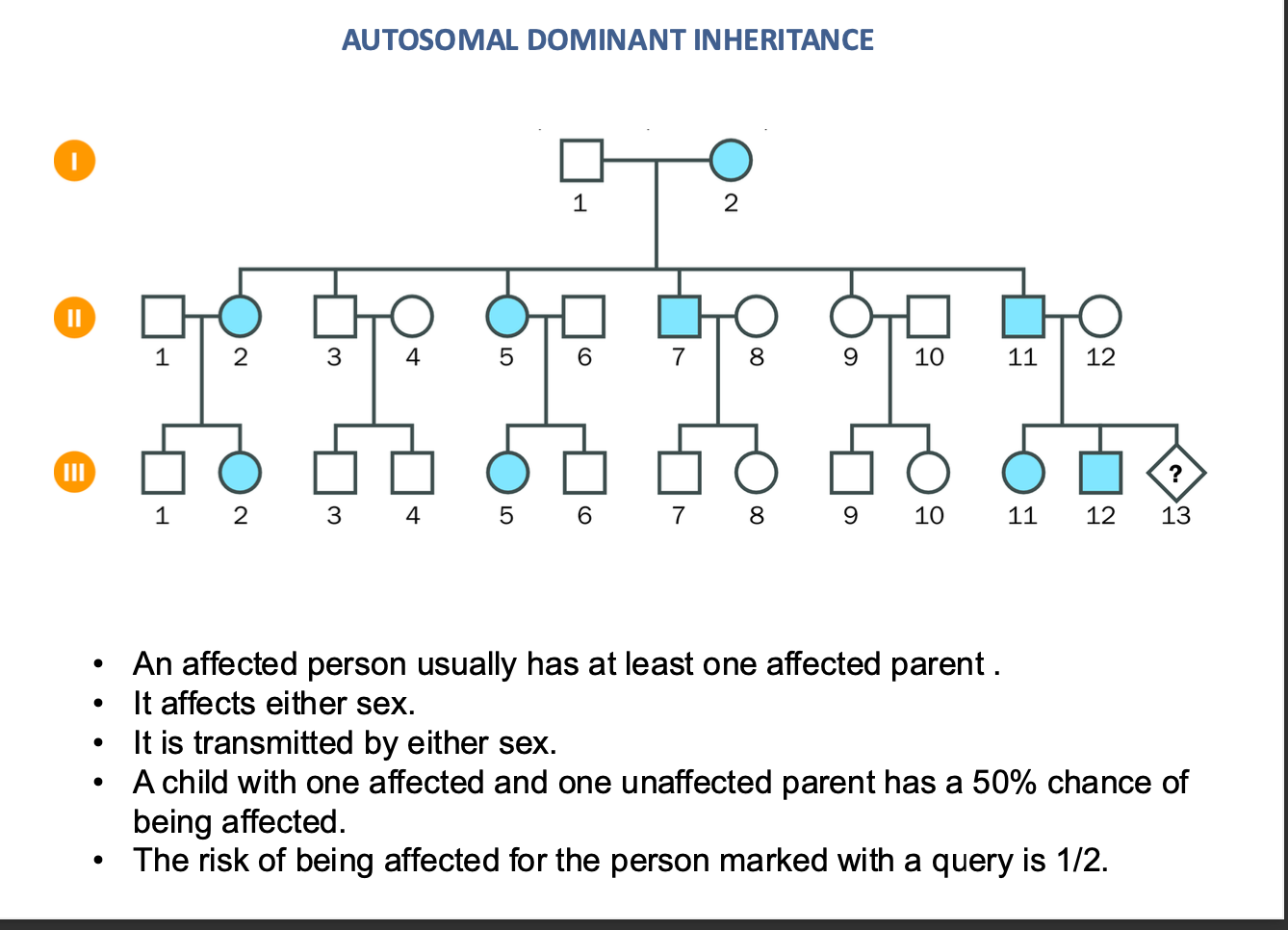
autosomal dominant inheritance
NOT AFFECTED BY SEX (XX OR XY)
50% chance
an Affected person will usually have an affected parent
TRANSMITS by either sex
AUTO SOMAL DOMINANT INHERITANCE
Child one affected / one unaffected parent
-50% chance of being affected
½ chance of being affected = risk of being affected = 50%
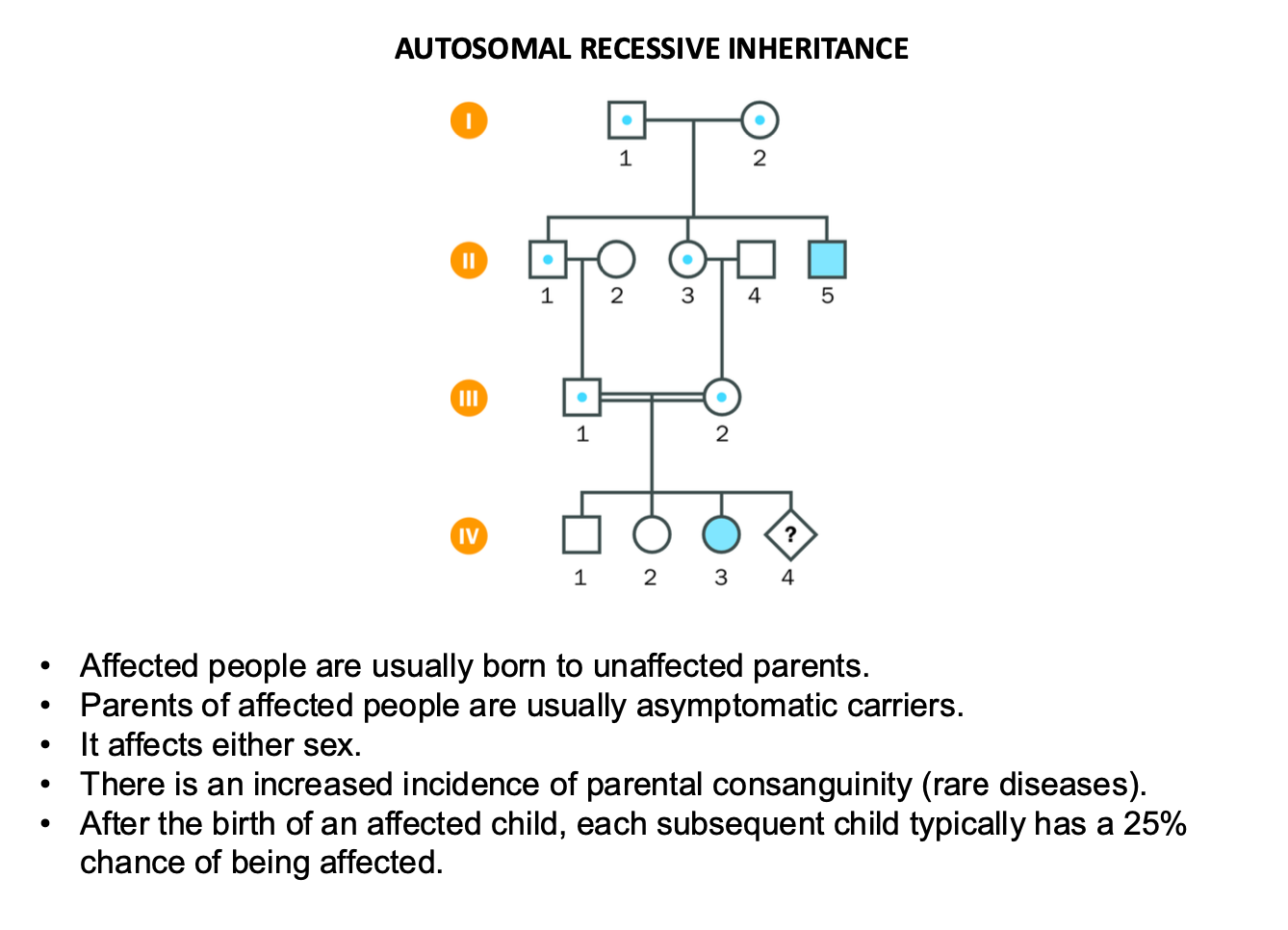
Autosomal recessive inheritance
Affected people usually born to unaffected parents
-Mum and Dad are carriers (blue dot)
-parents usually asymptomatic carriers
-affects either sex
-AFTER AFFECTED CHILD BORN
-Each child has 25% chance of being affected
X- linked recessive condition
Recessive - parents usaully not affected
-Affects mainly males
-Males usually affected have unaffected parents
-MOTHER USUALLY A CARRIER (asymptomatic)
Has affected male relatives
Can females be affected?
Yes - if father is affected + mother is a carrier
individual marked with ?
Would be 1 in 2 if male (MOTHER PASSES X, FATHER PASSES Y) MOTHER HAS 2 X so ½ chance they get her faulty chromsome
In recessive - female must inherit 2 FAULTY COPIES - one from mother and one from father (because females have 2 x chromosomes)
Carrier mothers have a 50% chance of passing the affected X chromosome to their sons, meaning the risk for an affected male child is 1 in 2 (50%).
x-linked dominant
1 parent IS AFFECTED
-Affects either sex but MORE FEMALES THAN MALES
-Females often more variably affected - due to having 2 x chromosomes
child of affected female - 50% chance of being affected
affected male - all his daughter but no son affected
(son not affected bc dad has disease and he passes y chromosome not the faulty x chromosome to his son)
Risk that individual marked with a query would be affect = 100% if female
0% if male (male passes y chromosome to son)
MALES PASS THEIR X CHROMOSOME TO THEIR DAUGHTERS
BUT ONLY THEIR Y CHROMOSOME TO THEIR SONS -
if father affected - the x chromosome passed to the daughter is affected
recap
recessive - usually parent not affected
recessive - daughter needs to get faulty gene from both parents
x-linked
Father GIVES HIS Y CHROMOSOME TO HIS SON
If father affected - boy will not be affected, girl WILL be affected
FATHER PASSES HIS ONLY X chromosome to his daughter
-the condition is DOMINANT which means only 1 is needed
mother (x,x)
father (x,y) - y to son, x to daughter (y linked diseases extremely rare)
Complications to basic patterns
Why?
name an: autosomal dominant + autosomal recessive disorder
Variable expression of individuals within the same family due to genetic background, modifier influences, environmental factors
Human characters showing dominant character can skip a generation due to variation in genetic background - nutrition, exercise and medication can lessen the severity (sometime)
sometimes can inherit a mild and aggressive version of the mutation in the same gene - can partially restore the function
VARIABLE GENETIC BACKGROUNDS
Waardenburg - autosomal dominant trait
Cystic fibrosis = autosomal recessive trait
Different family members show different features of type 1 waardenburg syndrome - an autosomal dominant trait
-although they have the same mutation in the PAX3 gene that encodes a transcription factor.
A UNIQUE GENETIC background can surpress the mutant allele (e.g. gene silencing) but the offspring can still inherit the mutant
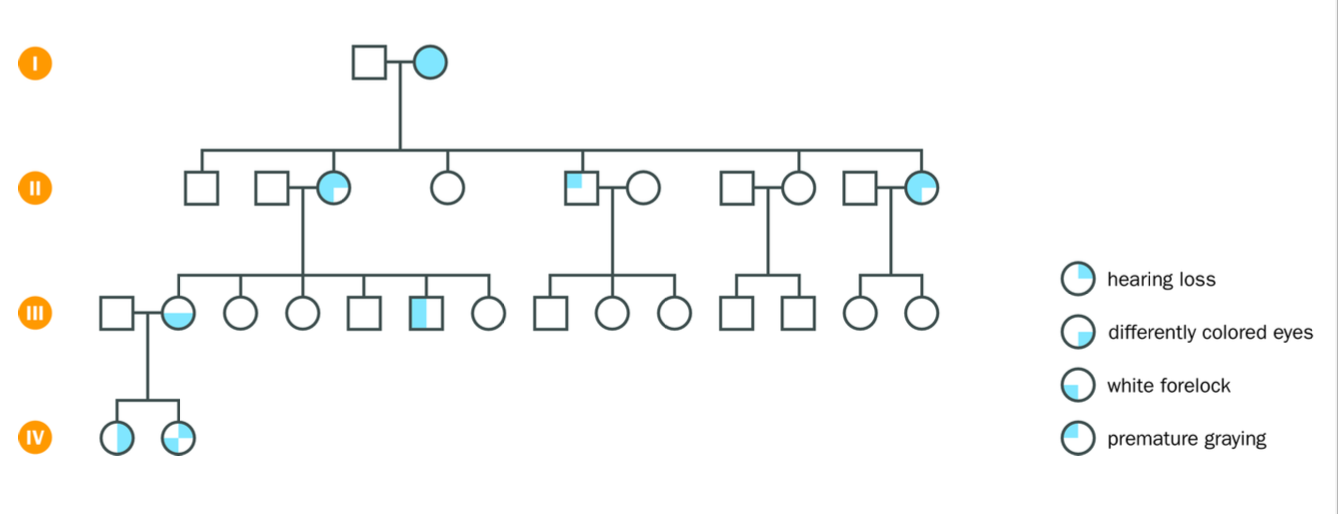
Complications to the basic patterns
-Discussed - genetic background causing variation in the expression of the faulty caused by the same mutation (waardenburg syndrome)- hearing loss, premature greying, white forelock, different coloured eyes
would expect 100% penetrance but…
sometimes due to random variation
extreme variable expression
dominant condition may fail to manifest itself
“skipping a generation”
variable faulty gene expression - due to genetic background influenaces
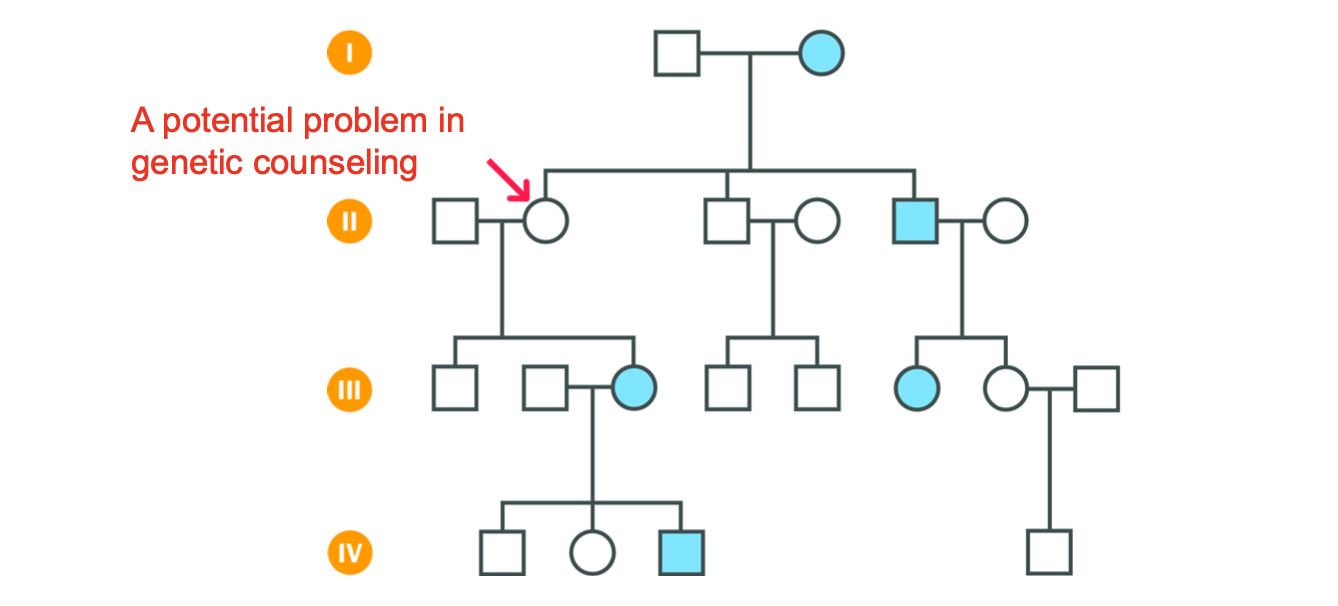
Age - related penetrance
Disease example?
-Huntington disease - neurodegenerative disorder
severity and probability of the symptoms depends on the age of an individual
Huntington proteins expressed in neurons in the brain - uncontrolled expansion - leads to the death of neurons and large gaps in the brain
A = probability of an individual carrying the disease allele developing symptoms by a given age
B = asymptomatic person who has an AFFECTED parent carries the disease allele
should undergo genetic testing to check if they carry the hd allele (HD allele will be present from birth)
even if an individual shows no symptoms, they may emerge with age.
neurodegeneration with age
Huntington gene protein expressed in neurons in the brain- glutamine in gene encoded by codon cagu - stretch of glutamine -
Copying of gene and replication and repair - CAGU can expand - above 36 copies - uncontrollable expanding Huntington protein - large stretches of glutamine
Protein precipitates - kill neurons - large gaps in the brain - removed neurons
Happens with age
Huntingtons disease = autosomal dominant inheritance (dominant usually has an affected parent)
50% chance of passing it down to each child ( not sex related)
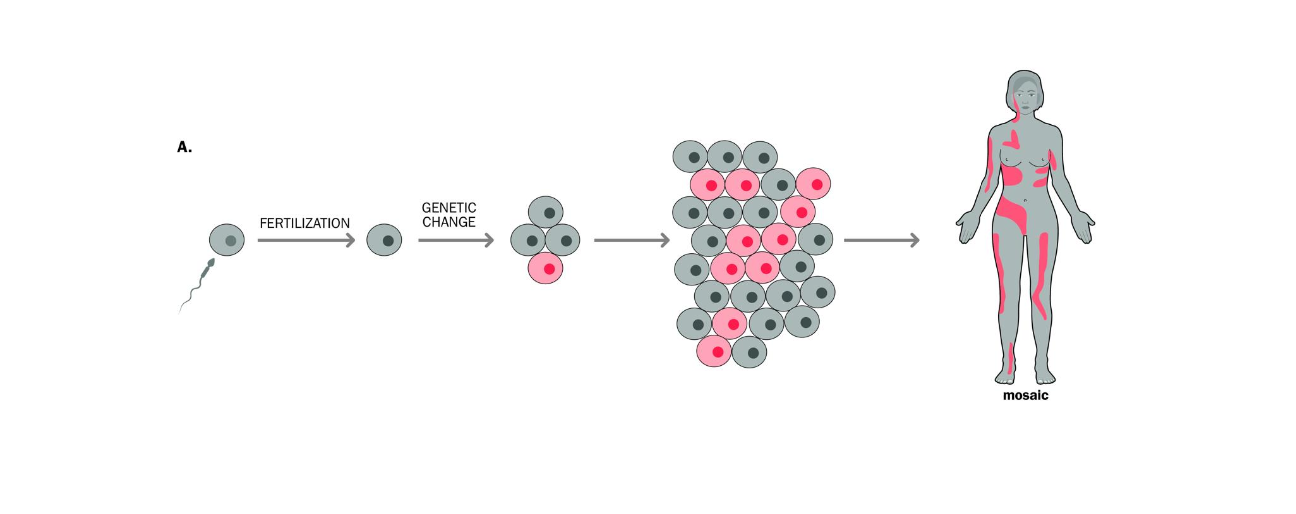
MOSAICISM AND NEW MUTATIONS
Mosaicism - Individual has 2 or more genetically different cell lines, derived from one original zygote
Post zygotic genetic change, meaning the mutations occurs AFTER fertilisation
Occurs
-Single cell is mutated after fertilisation
-Mutated cell DIVIDES -forms a population of cells genetically different from the rest of the cells in the body
straight AFTER FERTILISATION - mutated cells will contribute to a large number of tissues in the body -
If happens later not immediately after -mutated cells will contribute to a much lower number of tissues in the body (tissue - restricted mosacism) milder, localised manifestation)
IF MUTATION occurs EARLY IN DEVELOPMENT (AFTER fertilisation) - this affects a large number of individuals cells + contributes to a large number of tissues in the body..
The person may show signs of the mutation (phenotypic signs)
Later in development
Tissue-restricted - may not have the phenotype, or may have milder symptoms
Mosaic downsyndrome
-later stage mutation
some cells have the extra chromosome, some don’t - milder symptoms in the individual than if they had an extra chromosome in each cell
Another example is mosaic Huntington's disease, where the mutation in the HTT gene occurs later in life, and only certain tissues may show symptoms of the disease, leading to a milder or later onset of the disease.
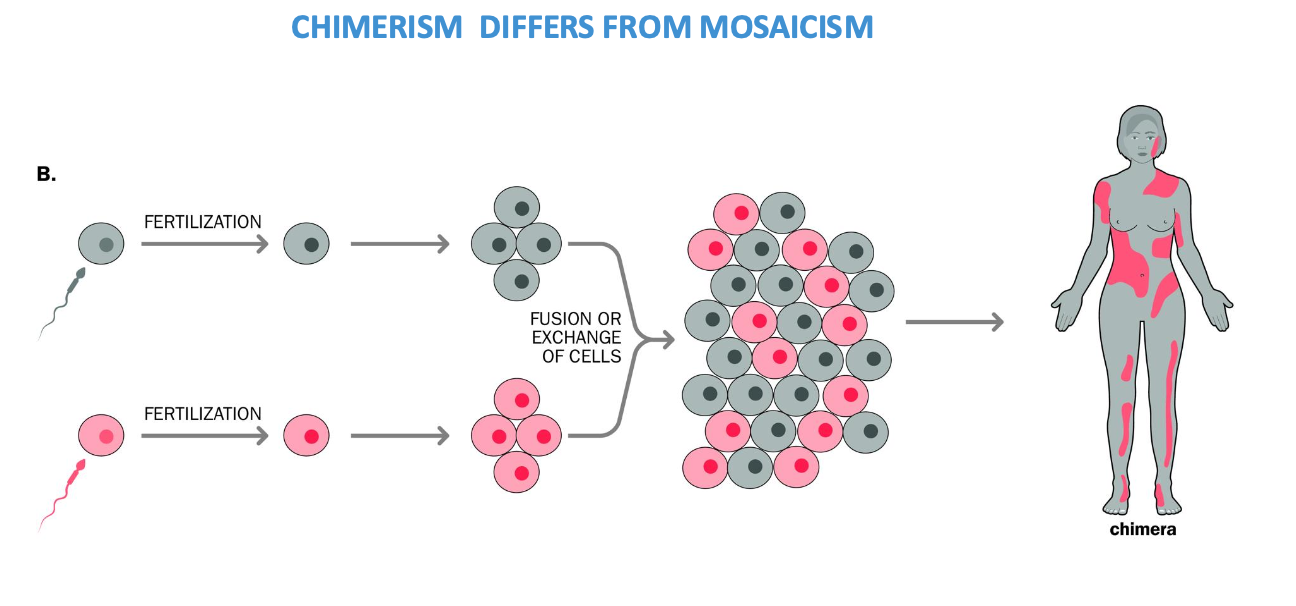
CHIMERISM
chimeras
Fusion of 2 zygotes to form a single embryo - form 2 genetically different cell lines
2 early stage embryos fuse during development
Often can have no visible impact on an individual
Chimerism - rare + proved by presence of too many parental alleles at several loci in a sample that is prepared from a large number of cells
Too many alleles in the same tissue on different loci - usually an individual inherits one allele from each parent
if testing a chimera - multiple tissues could show more than 2 alleles
New mutations are often present in … form?
what does this mean
COMPLICATING PEDIGREE INTERPRETATION
NEW MUTATIONS
(Genetic variation due to genetic background) another one
Parent may produce a single mutant gamete
post-zygotic expression - chimeras / mosaicsm
Often present in mosiac form
-This means they werent inherited from parents
-Mutation occured during the development of the embryo - long after or shortly after fertilisation
transmitted mutation may have occured
-Mutation in the germline ( cells that contribute to sperm / eggs) - it can be passed to offspring
mutation in non germline cells - wont be passed to offspring
PROBLEMS WITH SIMPLE PEDIGREE representation
Variation in genetic expression
age-related penetrance
new mutations
studying human chromosomes
methods:
Array Comparative genomic hybridisation (CGH)
Whole genome sequencing will replace all these methods
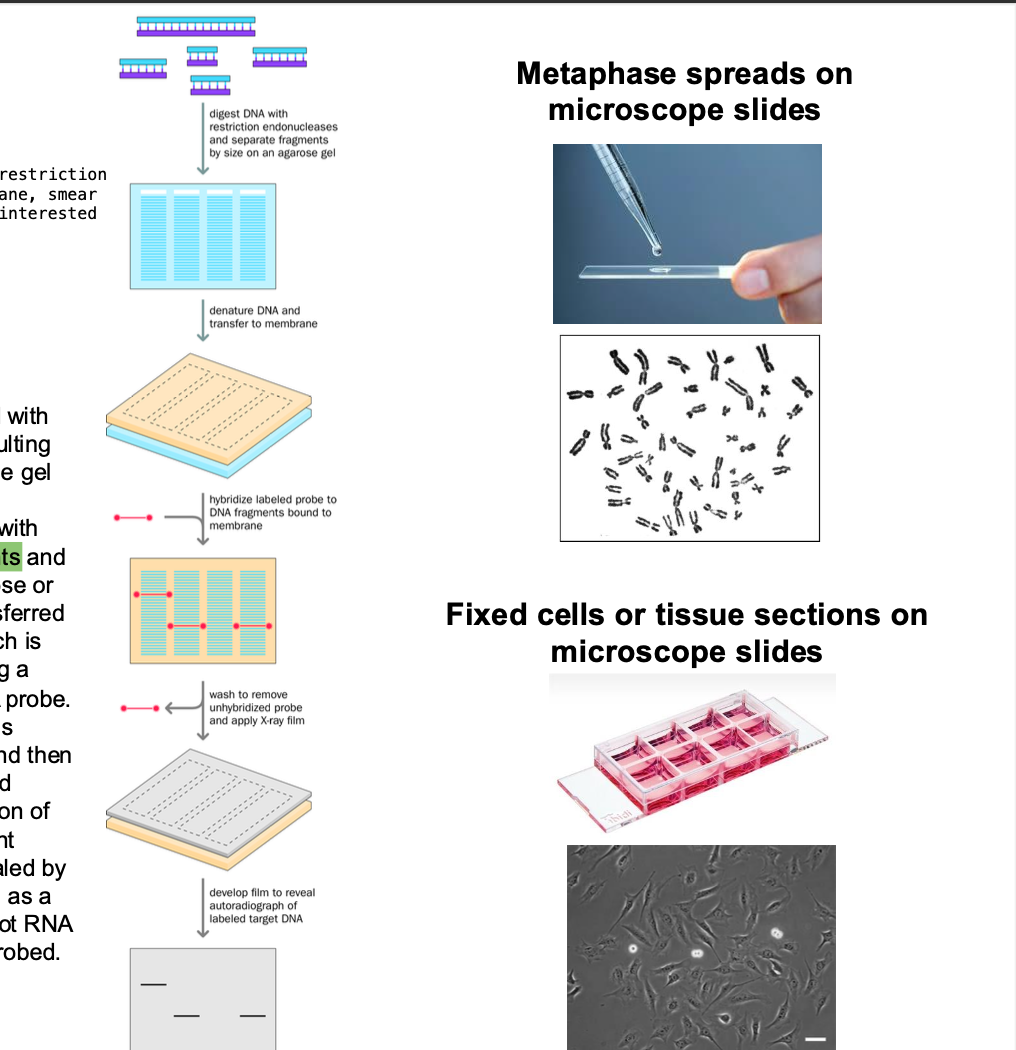
Karyotyping under the microscope
preparing a metaphase spread
-What is an important step?
Preparing metaphase spreads
-Disrupt the mitotic spindle using a microtubule poison such as colchicine
-Pauses the cell in metaphase
prevents aligning along the metaphase plate to progress to anaphase
-Colchicine inhibits microtubule polymerization - binds to tubulin blocking mitosis
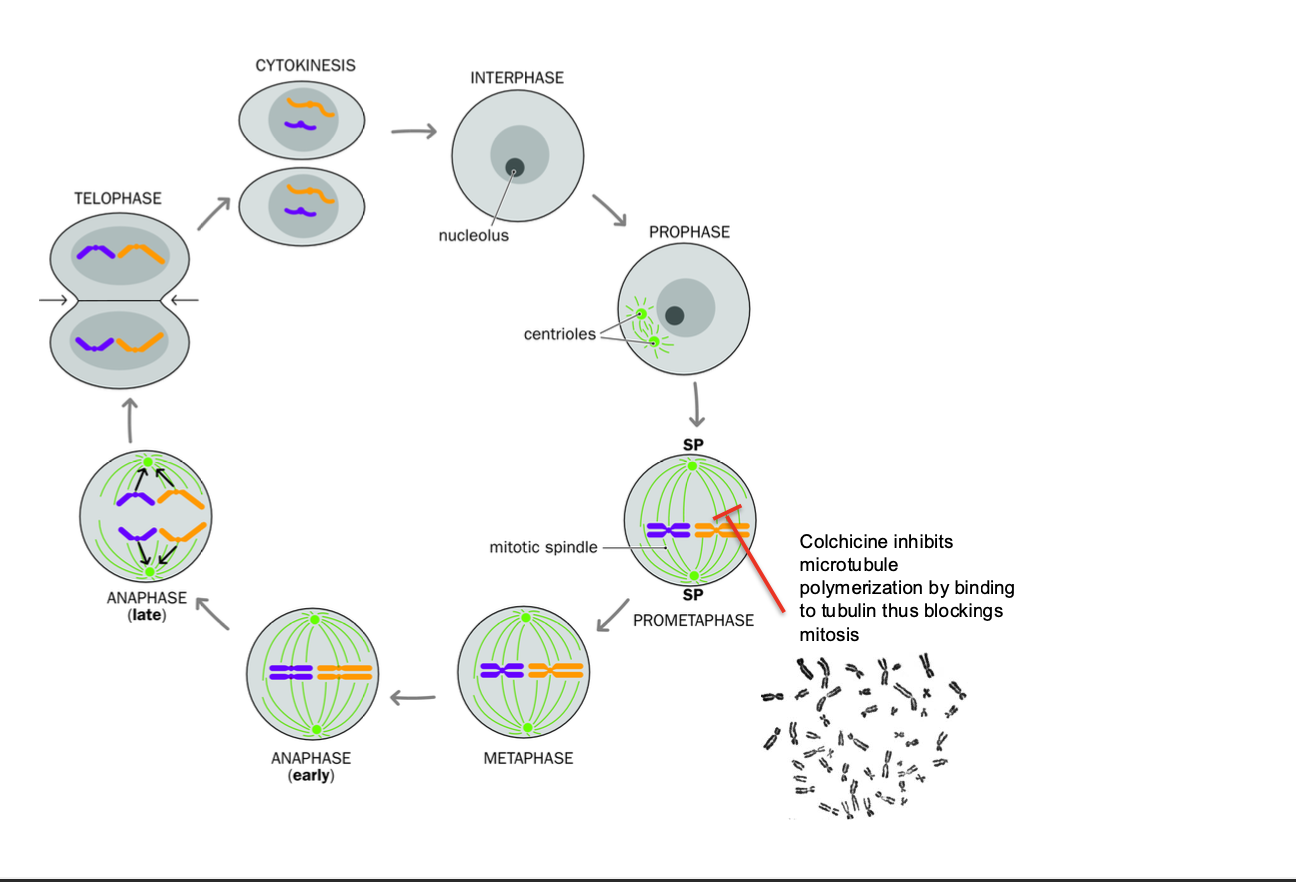
Preparation of metaphase spreads
Karyotyping
how are chromosomes identified?
metaphase - easiest to analyse for karyotyping
-We want to create a full set of chromosomes
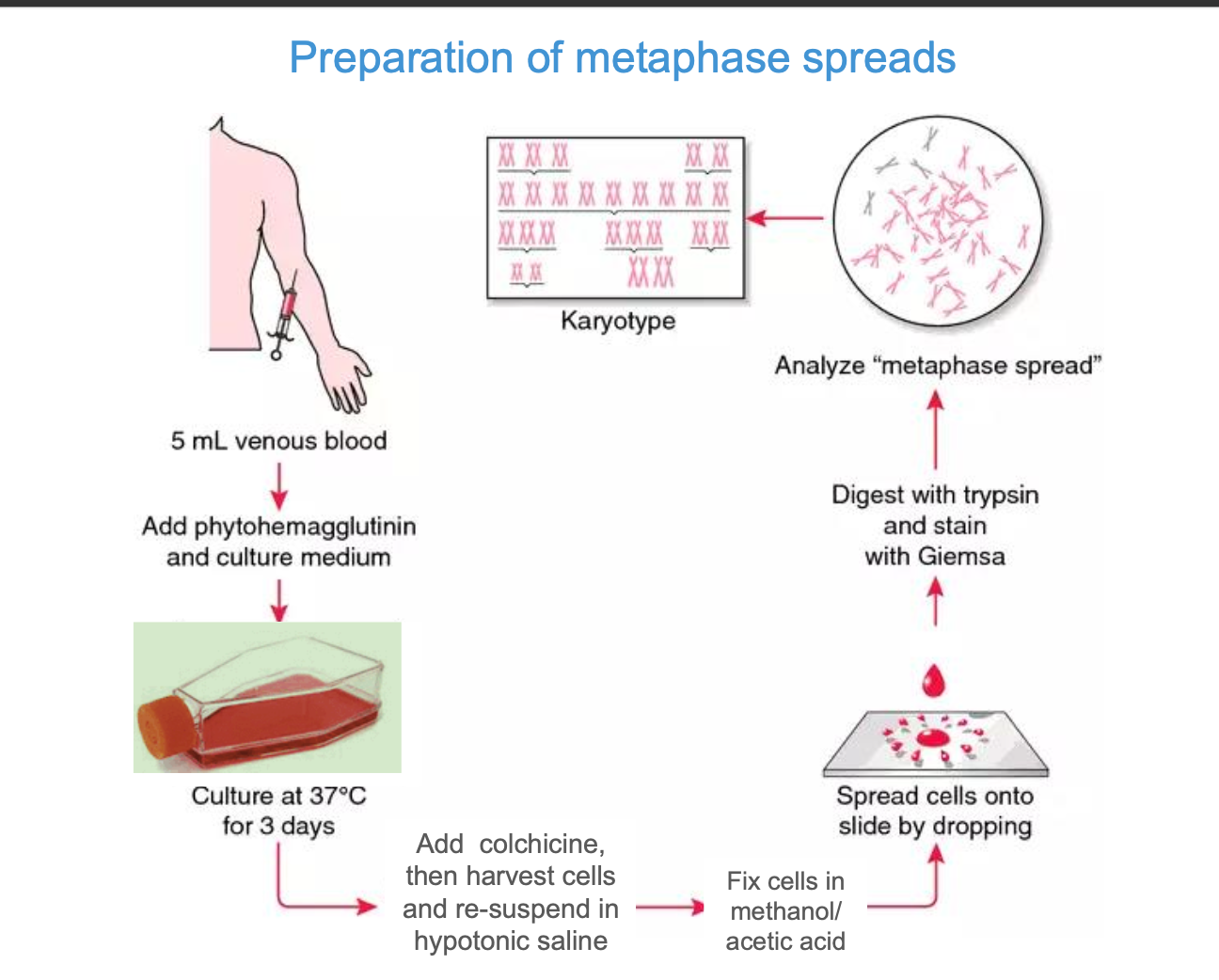
how?
Using a blood sample - extract 5ml of venous blood (want white blood cells, specifically lymphocytes - easily cultured + have a nucleus
extract 5ml venous blood sample
Add phytohemagglutinin (induces mitosis for clear karyotyping) and Sample is placed in a culture medium at 37 degrees celcius for 3 days
Add colchicine + harvest the cells and re-suspend in hypotonic saline - water enters the cells by osmosis - cells swell and are easy to visualise for chromosome analysis
fix cells in methano / acetic acid - preserves the chromosomes
digest with trypsin and stain with GIEMSA
-trypsin digests proteins on the chromosome surface such a histones - make the DNA easier to stain
Giemsa - binds to dna - creates a distinct pattern of light and dark bands along the chromosomes
Trypsin - controlled digestion
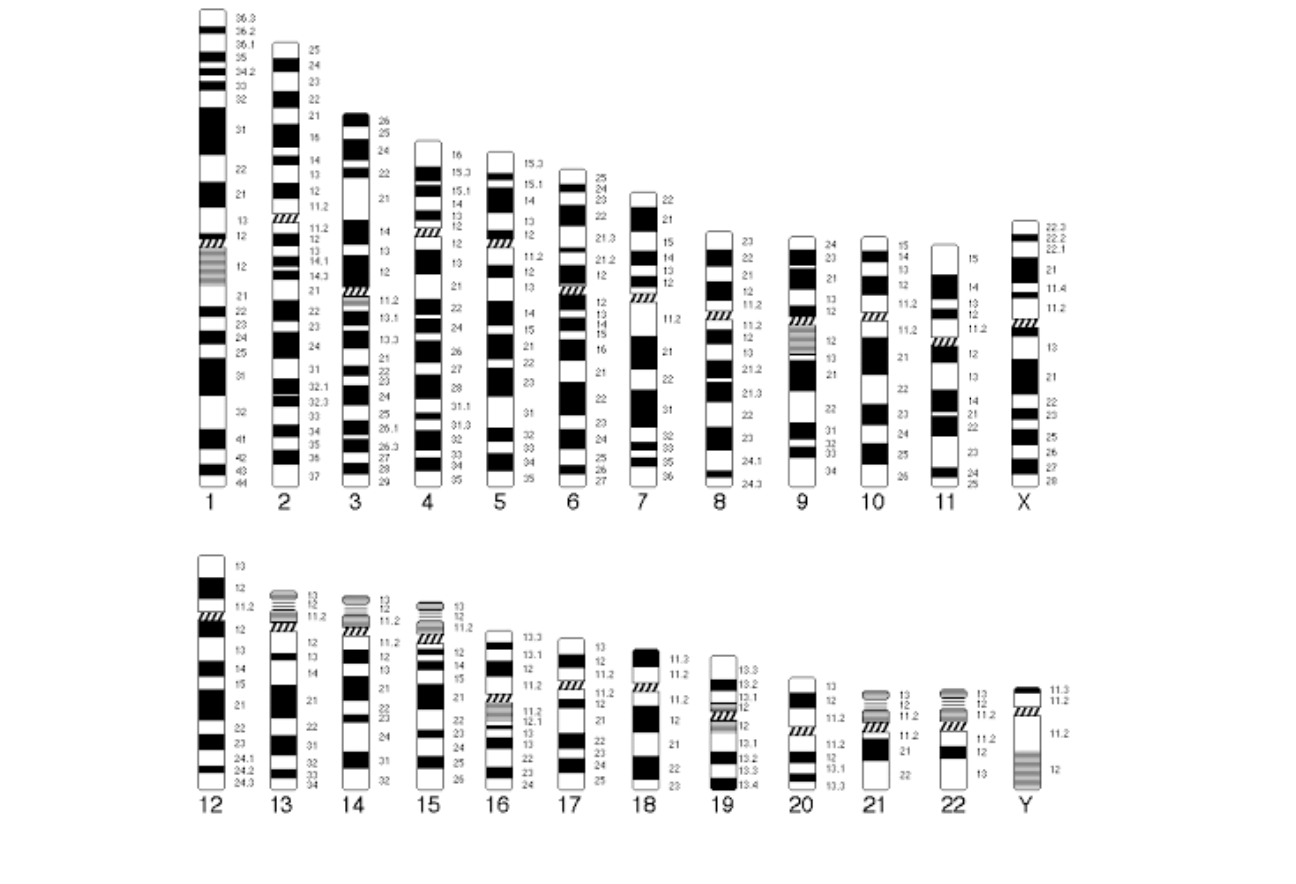
Identifying chromosomes
-identified by SIZE + BANDING PATTERN
Positively staining bands are known as G-BANDS (think g for Giemsa)
How are banding patterns displayed?
-banding seen depends on?
How are chromosomes grouped??
What is a satellite?
As chromosomal ideograms
-Banding seen depends on microscope resolution
different banding resolutions to resolve bands
Grouping
-Autosomes are numbered from largest to smallest BUT
chromosomes 21 - slightly smaller than chromosome 22
-Metacentric - centronmere is near the middle
Submetacentric - centromere at or near the end (think submarine under water)
Satelite - small segement separated by NONCENTROMERIC CONSTRICTION from the rest of a chromosome
-These occur on the SHORT ARMS of most acrocentric human chromosomes
Fundamentals of chromosomes mitosis + meiosis (genetic recombo)
Patterns + inheritance - identifying and studying chromosomes (metaphase spread) protection for genetic transfer
Chromsomal abnormalities - lecture 3
LECTURE 3
Chromosomal abnormalities
2 types
Polyploidy and aneuploidy
Structural chromosome abnormalities
Polyploidy
2 types
Triploidy - 3 full sets of chromosomes -fertilisation error
triploid embryo
1-3% of human pregnancies
-triploid embryo
-Rarely survive to term - not compatible with life (embryos)
error in fertilisation
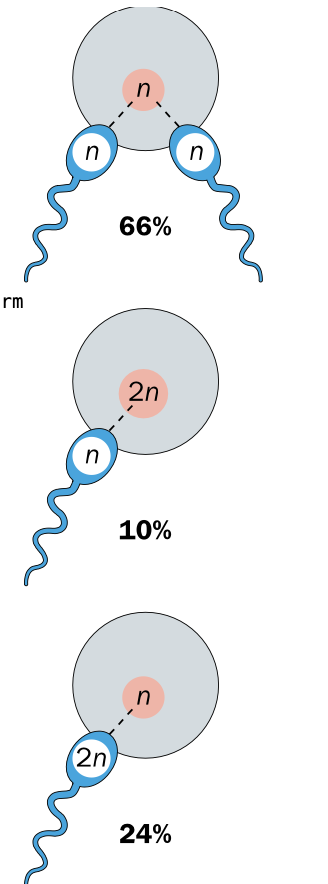
Dispermy - single egg fertilised by 2 sperm - accounts for 66% of triploidy
Diploid ovum - diploid egg fertilised by normal sperm 10%
Diploid sperm 24% - Diploid sperm fertilises normal egg
-Diploid = incorrect meiosis occured
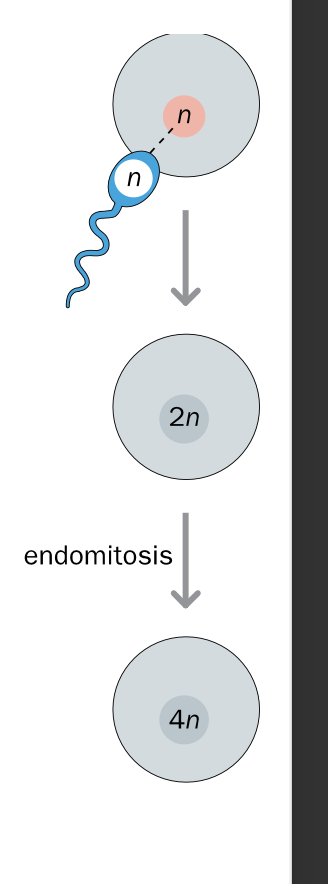
Tetraploidy - 4 full sets of chromosomes - ENDOMITOSIS cell division error
Normal fertilisation + fusion of gametes
AFTER FERTILISATION -incorrect cell division
NORMAL ZYGOTE
-Endomitosis - DNA REPLICATES without subsequent cell division
dna replicates BUT CELLS FAIL TO DIVIDE
Leads to ONE LARGE CELL WITH TWICE THE Normal amount of DNA
Lethal - result in miscarriage
Rare mosaic form - some tetraploid some normal diploid cells
-Survival still rare
Aneuploidy vs Euploidy
What is the cause of Aneuploidy?
Euploidy = having complete chromosome sets
polyploidy = having multiple chromosome sets
Aneuploidy - one or more individual chromosomes are present as an extra copy or are missing
-Abnormal number of chromosomes that is not a multiple of the haploid number
Aneuploidy - cause = nondisjunction / anaphase lag
(errors in chromsome separation during meosis / mitosis
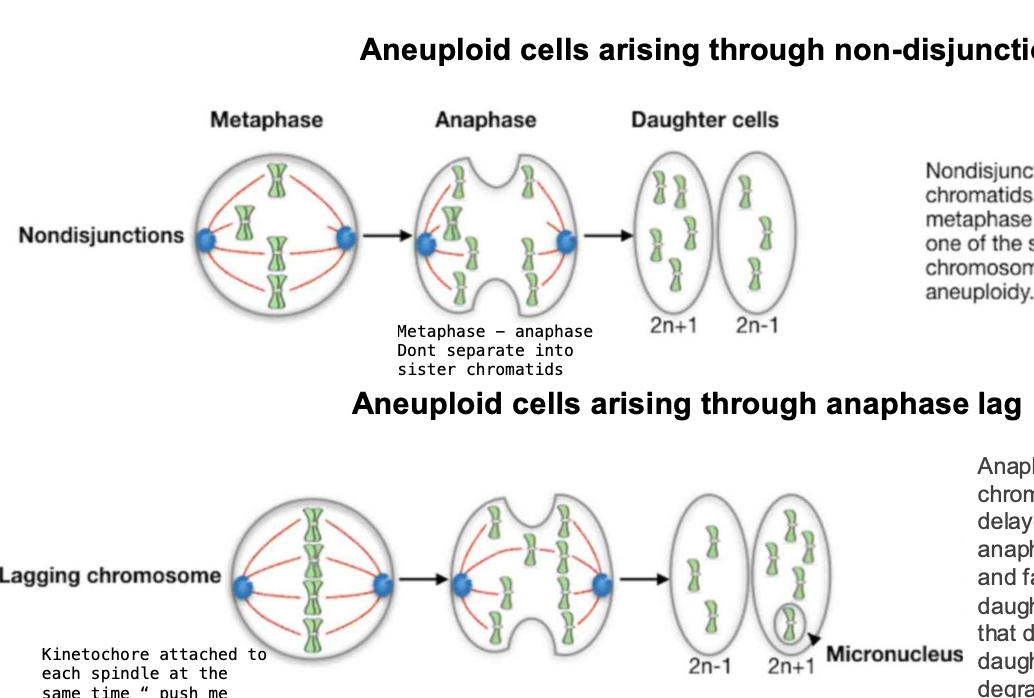
How do aneuploid cells arise
Explain
-Usually through nondisjunction in maternal meiosis
Failure of homologous chromosomes to separate properly during cell division
3 FORMS OF NONDISJUNCTION
(instead of separation of chromosomes - each go to the same cell and become fertilised)
Failure of a pair of homologous chromosomes to separate in meosis
failure of sister chromatids to separate during meosis II
Failure of sister chromatids to separate during mitosis
Nondisjunction during meiosis
produces gametes with 22 / 24 chromosomes
-Reasons not very clear - most cases due to maternal meiosis
Nondisjunction
produces one MONOSOMIC cell and one TRISOMIC daughter cell
Monosomic cell usually dies
-Trisomic cell can survive and establish MOSAIC triosomy
-Nondisjunction
-Anaphase lag
Aneuploidy
ANEUPLOIDY
-misalignment / lagging
Misalignment
Sister chromatids FAIL to align with the metaphase plate
-Remain at one of the spindle poles causing aneuploidy
-BOTH chromatids can enter ONE DAUGHTER CELL
trisomy / monosomy
Anaphase lag
-When a chromosome / chromatid is DELAYED in its movement during anaphase
-Lags behind the others + fails to be incorporated into the daughter nucleus
-Chromosomes that do not enter the nucleus of a daughter cell are eventually DEGRADED
or SOMETIMES persist in micronuclei as small membrane - bound vesicles
ANAPHASE LAG MECHANISM
-involves Merotelic attachements
Instead of the kinetochore being attached to one spindle pole, it is attached to microtubules from both spindle poles
-PUSH ME PULL ME EFFECT
-the chromosomes are pushed + pulled but dont move
lagging - may be excluded from the daughter cell
Cliical consequences of Aneuploidy
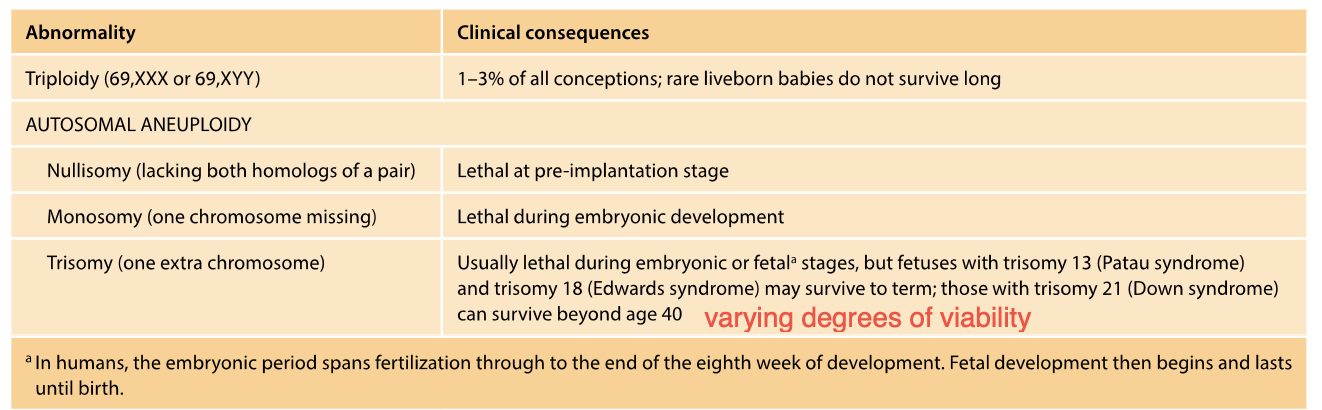
-TRISOMY
some survive to term? others dont? exception?
-MONOSOMY
-Usually lethal
-Multiple abnormalities present at birth (congenital)
-Trisomy - extra chromosome - even though the chromosome is perfectly normal - congenital implications (present at birth)
-Trisomy 13 and 18 - CAN SURVIVE TO TERM -sever malformations - incompatible with long term survival
Most autosomal trisomies - NOT COMPATIBLE WITH SURVIVAL
unless mosaic form (only some cells are affected, while others have the normal amount of chromosomes) - early development stages
MONOSOMY
Autosomal MONOSOMIES - EVEN MORE CONSEQUENCES - Lethal, catastrophic
ANEUPLOIDY - Imbalance of different chromosomes and genes e.g. transcription factors
monsomies - MORE PROFOUND EFFECT
-one missing worse than having 3
DOWN SYNDROME IS A TRISOMY OF CHROMOSOME 21
EXTRA SEX CHROMOSOME (STRAYING FROM AUTOSOMES
In females - random inactivation
FAR FEWER ill effects
-Can function normally or have relatively minor problems compared to AUTOSOMAL TRISOMY - 47 xxx 47 XXY - can function normally
Monosomy in sex chromosomes - fewer consequences
45,X - turner syndrome
turner syndrome
45 Y is lethal - y chromosome is essential for male development BUT does not contain the genes required for survival (located in the x chromosome)
-short, infertile, learning disabilities, hearing loss etc.
Random inactivation of X CHROMOSOME IN FEMALES
In females, (XX) one of the X chromosomes is randomly inactivated - One x chromosome remains active per cell
Ensures, like men, women have a similar x-linked expression (men have 1 x chromosome and ONE Y)
-TURNER SYNDROME
One X chromosome is missing - the single x remains active in cells - no x inactivation occurs.
x- chromosomes - contain ESSENTIAL GENES
AUTOSOMAL CLINICAL COSEQUENCES ARE WORSE THAN SEX CHROMOSOMES

How do CHROMOSOME STRUCTURAL VARIANTS EMERGE?
-Mis repair of damage -especially DNA DOUBLE STRAND BREAKS, can be recombination errors / replication errors.
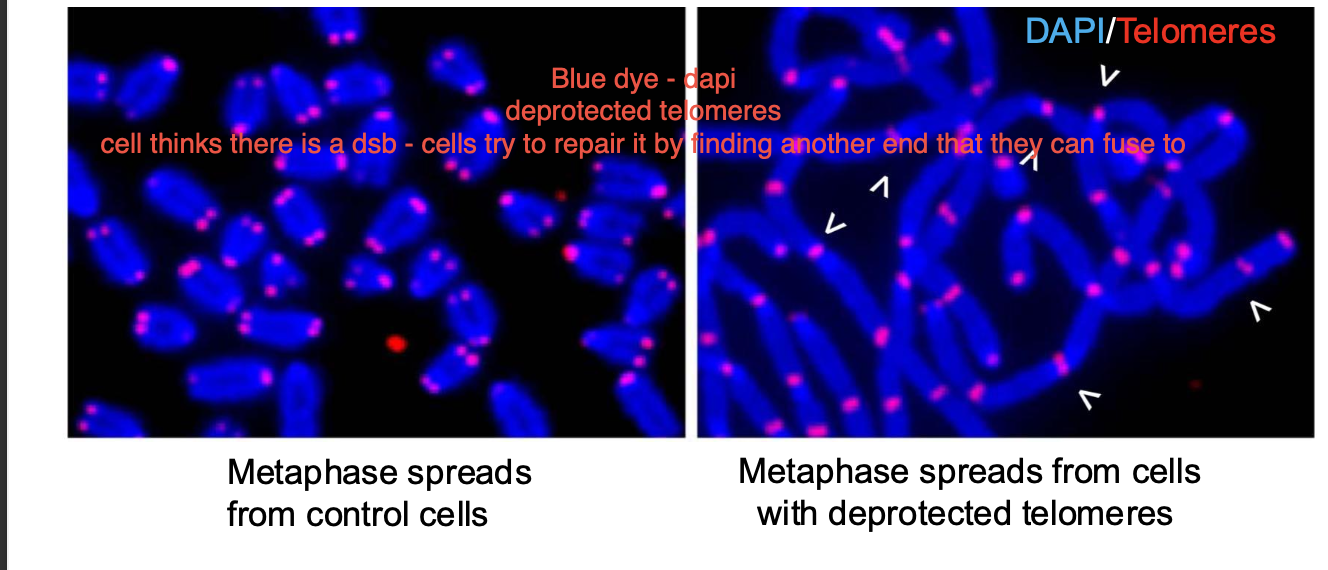
Shelterin - telomerase protection the ends of chromosomes - prevents them from being recognised as a double strand break - prevent INAPPROPRIATE DNA damage response. (deletions, duplications, inversions) inappropriate recombination - during meiosis
Without telomeres - ends of chromosomes are TREATED as double strand breaks
telomeres - crucial protection + stability of the chromosome
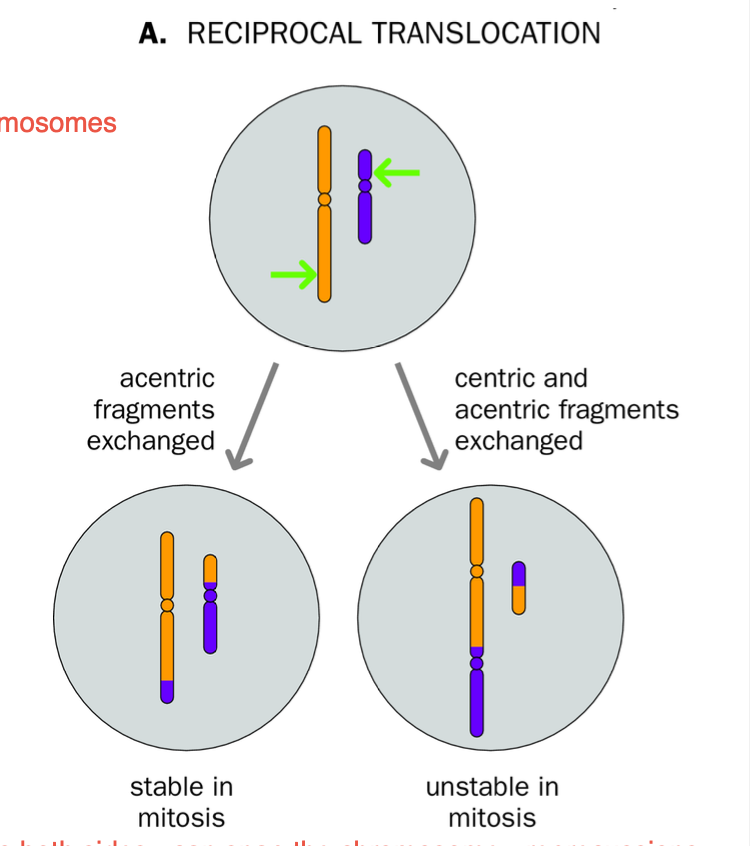
Reciprocal TRANSLOCATIONS
-Structural variants of Chromosomes
EXTENT OF CONSEQUENCE OF RECIPROCAL TRANSLOCATIONS
DEPENDS ON?
Mis repair of double strand breaks
-inappropriate recombination (during meiosis) (remember this is where recombination occurs) - obviously recombo problems cause variation
Errors in replication
Problem
-More than 2 BROKEN ENDS - repair machinery JOINS THEM TO THE WRONG PARTNERS.
2 nonhomologous chromosomes swap segments.
-CHROMOSOMES ARE A BLEND OF THE ORIGINAL CHROMOSOMES
-if each chromosome variant still has ONLY ONE CENTROMERE - then MITOSIS CAN OCCUR WITH NO PROBLEMS
DICENTRIC CHROMOSOME
-Occurs when 2 parts of a chromosomes swap and both RETAIN THEIR CENTROMERE.
Chromosome now has 2 centromeres
-mitotic spindle responsible for chromosome separation - struggles to pull chromosome to opposite poles of the cell
Mitotic spindle CANT DEAL with a chromosome with 2 centromeres.
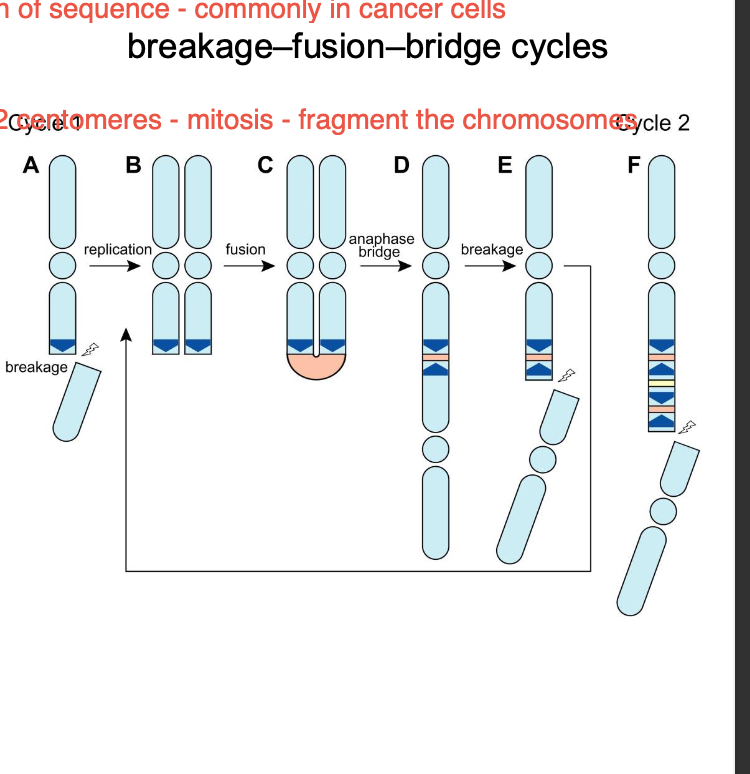
Breakage - fusion - bridge cycles - MITOSIS OF DICENTRIC CHROMOSOME
-Tension, spindle will try to pull chromosome in two directions at once
Breakage - double stranded break of chromosome
Fusion - non homologous during Reciprocal Translocation
Bridging - during cell division
breakage - separation
Break fusion break - cycle occurs where the segments of the chromosome continuously
Separation of chromosomes fails to successfully occur - ANEUPLOIDY
Cause fragmentation + genomic instability
-HALL MARK OF CANCER IN CELLS - CHROMOSOMAL ABNORMALITY
ACENTRIC CHROMOSOME - ANEUPLOIDY
Chromosome has LOST its centromere due to translocation
-Chromosome CANT be pulled to either pole during mitosis
ACENTRIC CHROMOSOME
-Usually lost during mitosis - fails to segregate into daughter cells due to MISSING CENTROMERE
-Results in genetic loss + instability
ANEUPLOIDY
MITOSIS -
Acentric - loss of genetic material
Dicentric - bridge - fusion - break cycle - MITOTIC SPINDLE CANT HANDLE 2 CENTROMERES - fragmentation of chromosomes
genetic INSTABILITY
Reciprocal Translocation scenarios
-Dependent on?
Another> think breaks? multiple breaks (2)
LOSS OF TELOMERES>
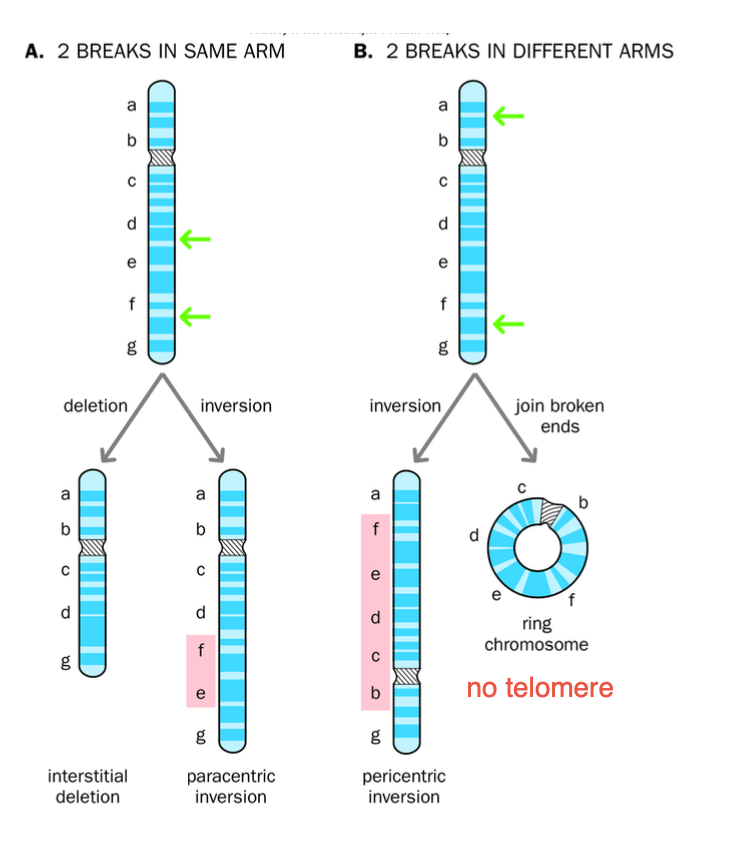
INVERSION = 2 BREAKS
Para = same arm
Peri = different arms
centromere
Dicentric - chromosome has 2 centromeres - (Joining of 2 segments) - mitosis - chromosome strain - bridge -fusion - break
Acentric - chromosome LOST its centromere - lost during meiosis- unable to separate in sister chromatids
2 BREAKS on one chromosome
-Incorrect repair of break + NO LOSS OF MATERIAL
-Paracentric / pericentric inversions
Paracentric - inverted segments DOES NOT contain the centromere
-Broken segment FLIPS but stays within the same arm of the chromosome
-No change in genetic information, BUT THE ORDER OF THE GENES IS CHANGED
-Can cause problems during mitosis - IF CROSS OVERS occur - dicentric / acentric chromosomes may arris
Pericentric - inverted segment contains the centromere
Inversion on different arms - centromere included
Gene order is altered, no genetic material is lost
Crossing over leads to DUPLICATIONS AND DELETIONS IN CHROMOSOMES
-misalignment during meiosis
Unbalanced chromosomal rearrangements in offspring
Congenital abnormalities
-GAMETES fail to fertilise
INVERSION RESULTS IN 2 BREAKS
Paracentric - breaks are on the same arm
Pericentric - breaks are on different arms.
Structural variants
Mis-repair of breakages
-Centromeres - acentric / dicentric - para / peri (arms / centromeres included)
Loss of telomeres - formation of RING CHROMOSOME
-Ends of chromosome fuse together
-Loss by EXTREME SHORTENING
Associated with intellectual disabilities, growth delays + congenital effects
Multiple breaks on a single chromosome
-No loss of material?
names
INVERSIONS
Breakage then inversion
Paracentric - breaks are on the same arm
Then inversion occurs
-Pericentric -breaks are on different arms
Inversion then occurs
The order of the gene is altered- due to inversion
BUT no genetic loss
Loss of telomere / no telomere
ends fuse together - create a ring chromosome
(same arm breakages ) - can produce acentric chromosomes
Repair can produce an acentric fragment - LOST DURING NEXT MITOSIS (missing chromosome) - genetic instability
Interstitial deletion / ring chromosome - fusion of chromosome breakages in a circular manner - absence of telomeres
interstitial genetic loss - segments between the BREAKAGES unable to reattach
ribosomal gene arrays are found on?
-Function of ribosome gene arrays
RNA ONLY FOUND ON P ARMS
p arms - short + consist of repetitive DNA
Only found on P ARMS of acrocentric chromosomes
-CENTROMERE is located AT THE END
Function:
Protein synthesis
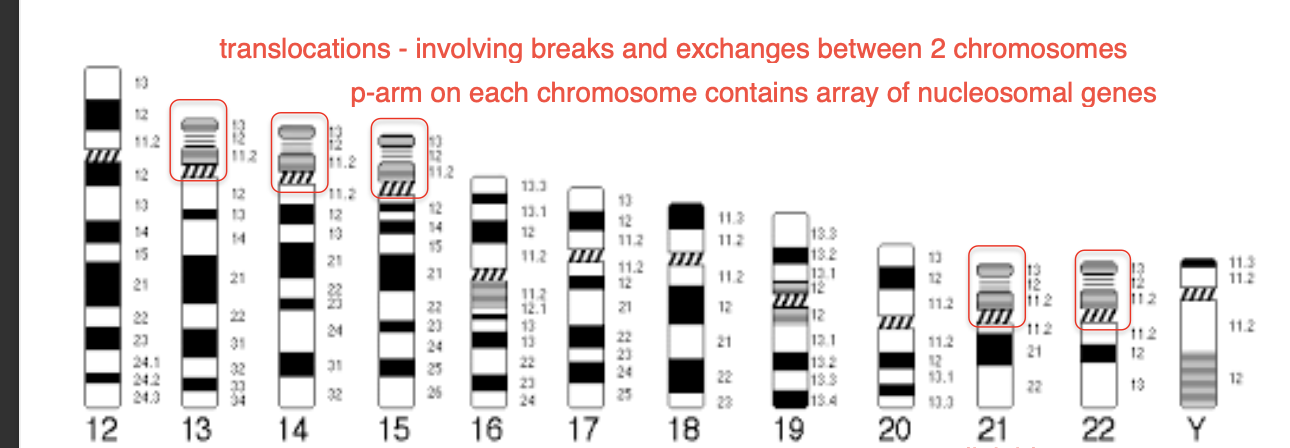
Robertsonian translocations
-Joins 2 acrocentric chromosomes - 13,14,15,21,22
-MOST COMMON FORM OF HUMAN TRANSLOCATION
acro-centric chromosomes
-Very small
Largely repetitive ribosomal RNA genes rRNA genes
SAME sequence found in SIMILAR BUT NOT IDENTICAL CHROMOSOMES (ACROCENTRIC)
-RECOMBINATION between homologous sequences in HETEROLOGOUS short arms (not identical but evolutionarily similar origin)
=Acentric - missing centromere
Dicentric products - 2 centromeres one from each chromosome fused together
USUALLY
Two centromeres of the dicentric chromosome are SUFFICIENTLY close together - function as a single large centromere
-1 CENTROMERE IS EPIGENTICALLY SILENCED
-FUSION chromosome SEGREGATES REGULARALY + STABLE IN MITOSIS
-prevents genomic instability + PREVENTS break- fusion - bridge cycles
Acentric fragment- LOST
genetic information is lost - doesnt affect PHENOTYPE
as rDNA repeats are found in high numbers in all other acrocentric chromosomes
PRESENT IN EVERY 1 IN 1,000 BIRTHS
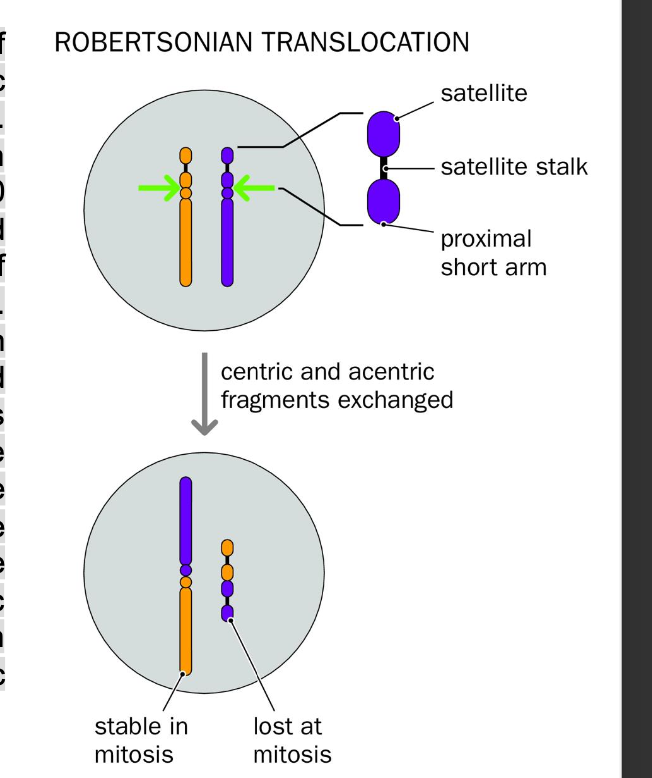
Consequences of structural chromosome abnormalities
are robertsonian considered balanced?
CAN BALANCED chromosome abnormalities still affect the phenotype?
-give an example of this
abnormalities
Structural variants - TRANSLOCATION (Reciprocal/Robertsonian
-BALANCED?
-No net gain / loss of chromosomal material
balanced - no phenotypic effect (usually)
Unbalanced
-May have an effect
Robertsonian- balanced - no phenotypic affect
-Considered balanced - no phenotypic effect
epigentically silenced centromere
lost centromere - acentric genetic info lost but rDNA is plentiful in other acrocentric chromosomes - loss of genetic material is negligible / redundant
SOME balanced abnormalities AFFECT THE PHENOTYPE
-impercise repair - deletion, duplication, mutation of IMPORTANT GENES
-chromosome break could disrupts an IMPORTANT GENE
-An active gene could be translocated to HETEROCHROMATIN where it is silenced
-Gene is separated from its enhancer
DYSTOPHAN - muscular dystrophy
-Breakage near the DMD gene causes the DMD gene to translocate to heterochromatin- silences the gene causing muscular dystrophy
Over expression? causes?
-Balanced abnormalities - malignant phenotype
C-MYC gene
over expression causes CANCER - GENE AMPLIFICATION
C-MYC GENE - CAUSES CANCER
-Over expression
-Translocation between chromosomes 8 and 14
C-myc gene is translocated/ juxtaposition to immunoglobulin HEAVY CHAIN LOCUS
-MYC overexpression
MCY is upregulated in B lymphocytes
-Malignant phenotype
Uncontrolled Cell proliferation
consequences of structural abnormalities in MEIOSIS
ABNORMALITIES WITH 1 CENTROMERE- balanced
-Transmission through mitosis NO PROBLEMS
-No excess / missing material
Abnormality = Balanced
Normal PHENOTYPE - usually
Exceptions - Muscular dystrophy + BURKITT’S lymphoma
Mitosis is fine
PROBLEMS ARISE IN MEIOSIS THOUGH - Homologous chromosome pairing
-Rearranged (variant) chromosomes DO NOT match their normal counter parts
-Homologous sequences between the chromosomes still pair
Unbalanced gametes
-excess / missing genetic material
Translocations - form CROSS STRUCTURES
Contortions of chromosomes
-BALANCED abnormalities
-No problem getting through mitosis
-Meiosis - cross structures / contorted chromosome structures as HOMOLOGOUS SEQUENCES join together but chromosomes as a whole are different due to variation (improper repairing - double strand breaks - cross over)
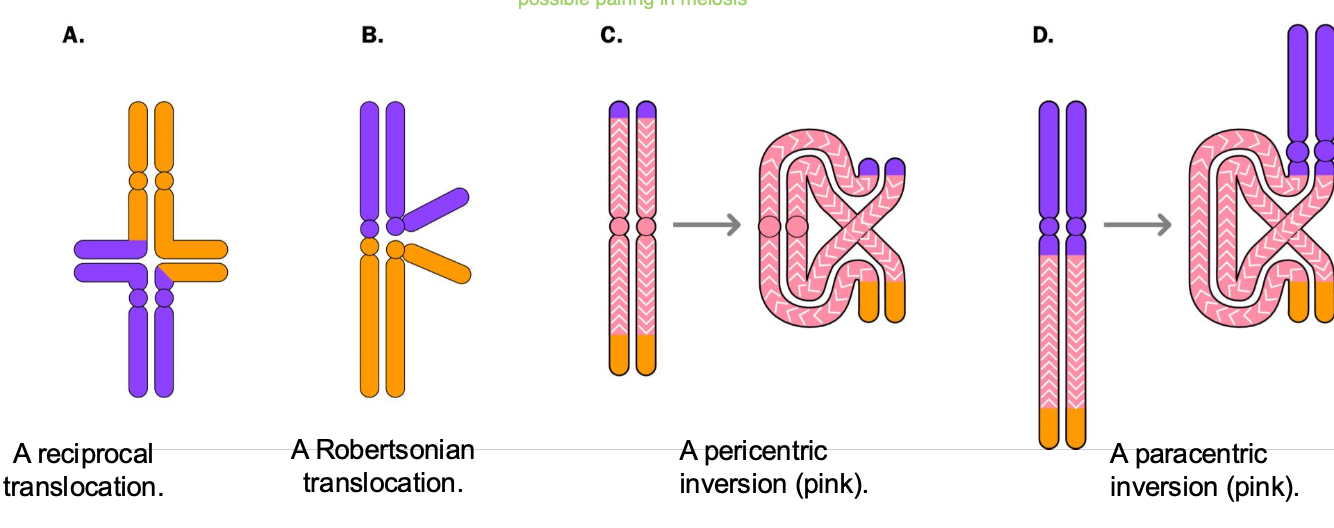
can be a carrier of translocation
can be normal
trisomy / monosomy - after fertilisation aneuploidy - maternal meiosis
Remember aneuploidy - disjunction / anaphase lag
-NON DISJUNCTION - leads to mono / trisomy
(remember polyploidy = additional chromosome sets)
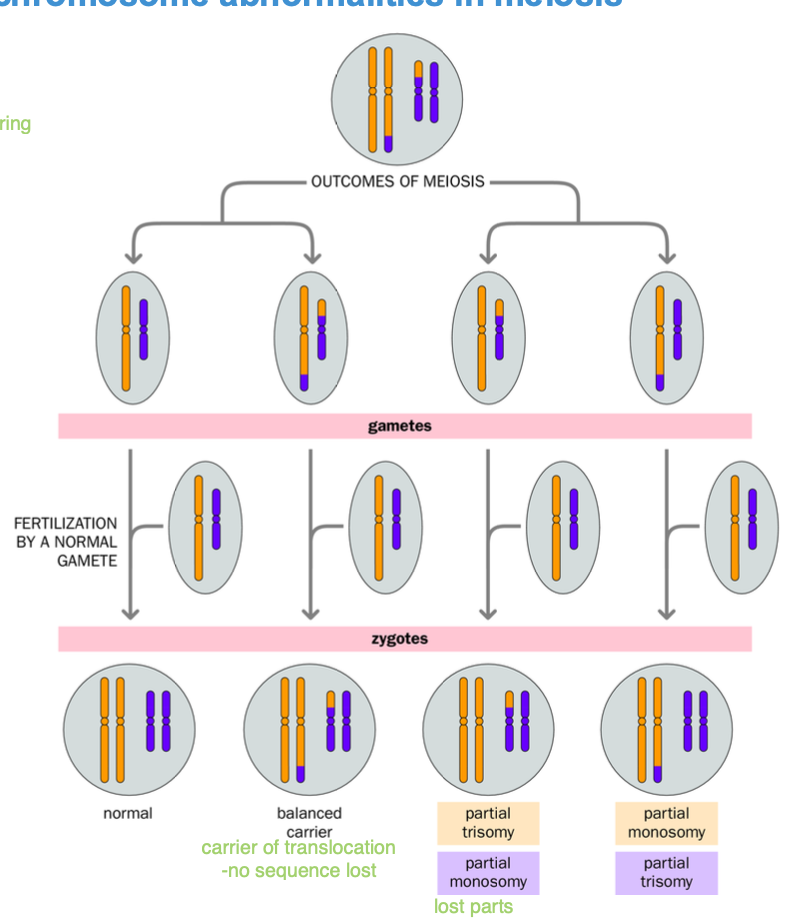
consequences of structural chromosomal abnormalities in meiosis
(remember meoisis is where the abnormality is detected BECAUSE chromosomes are not homologous to fuse together - so only fuse partially
-CAUSING ANEUPLOIDY - trisomy / monosomy
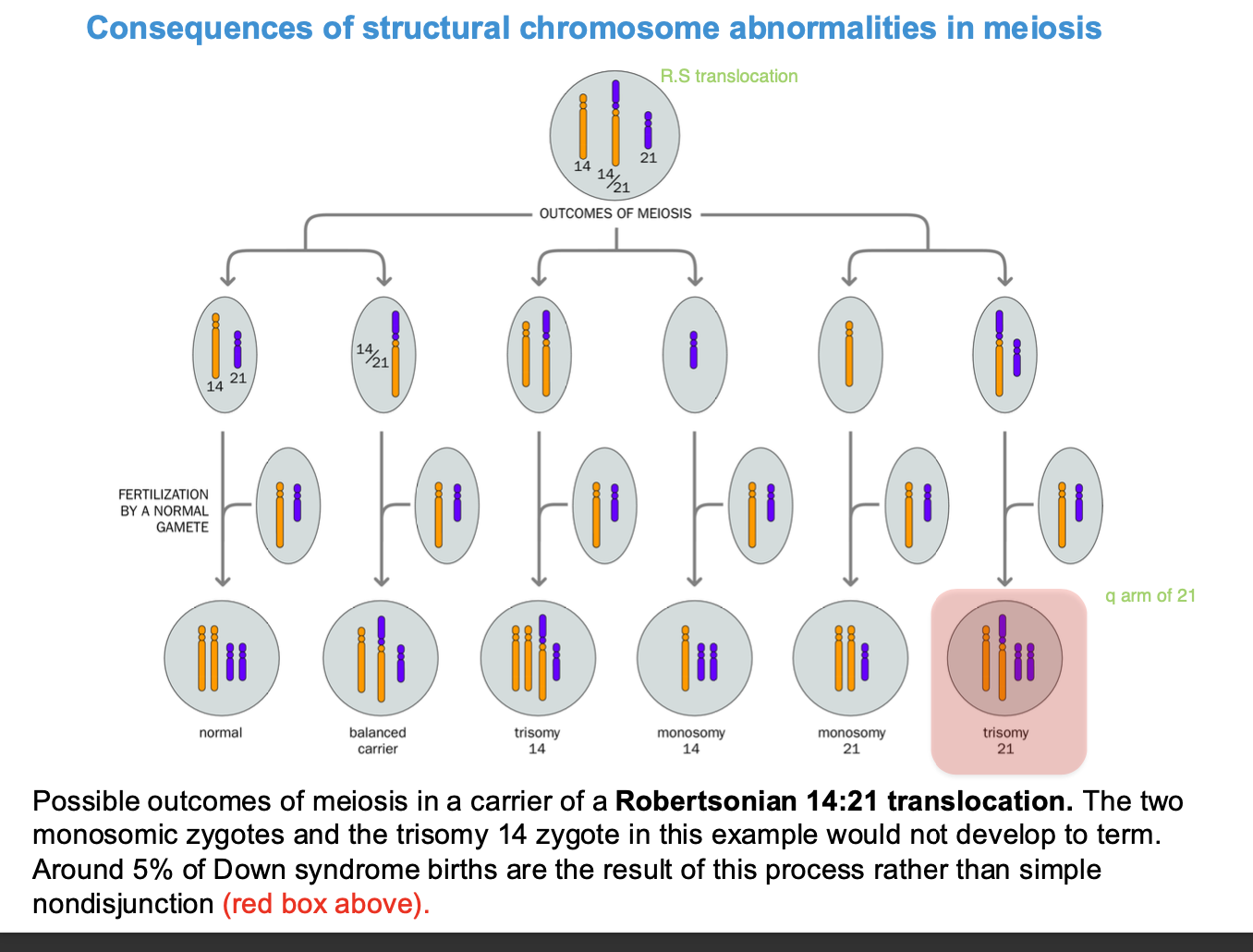
ROBERTSONIAN 14:21 - TRANSLOCATION
Robertsonian = acrocentric chromosomes
5% of downsyndrome - due to ROBERTSONIAN TRANSLOCATION
aneuploidy - chromosome 14 is translocated to chromosome 21
trisomy
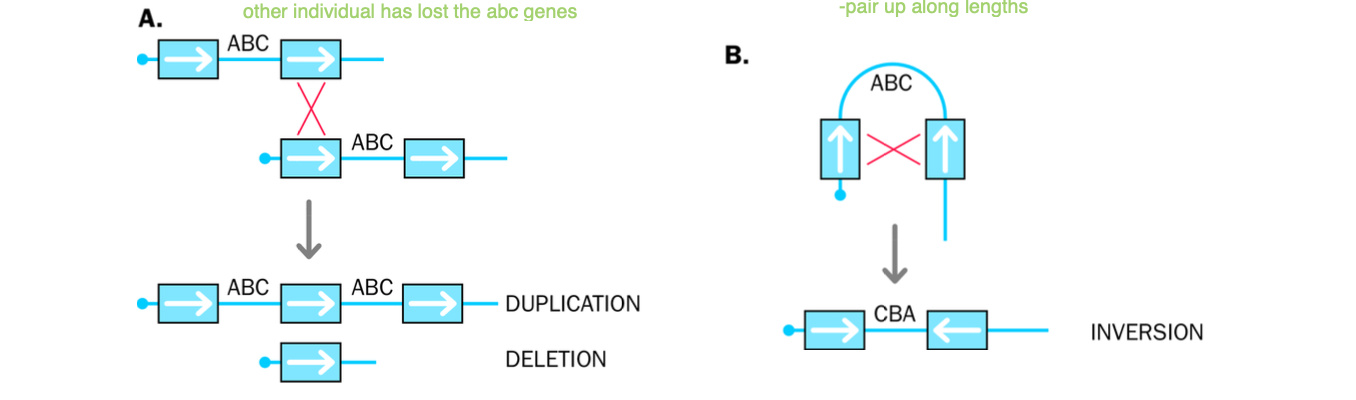
structural variants
-can be small duplication or deletions
can be large scale dna rearrangements either
Recurrent structural variants
NAHR
-non allelic homologous recombination (alternative forms of the same gene = allele)
OR UNEQUAL CROSS-OVER
-Occurs between 2 repeated sequences
-repeated sequences
-Meiosis - unequal cross over - deletion vs duplication
cross over - non homologous chromosomes EXCHANGE genetic information
unequal cross - over
-1 chromosome - recieves duplications of a region of the genome (duplication)
-1 chromosome loses the region of the genome (deletion)
MISALIGNMENT OF LOW COPY REPEATS
Unequal cross - over - NAHR - NON ALLELIC HOMOLOGOUS RECOMBINATION
LOW COPY REPEATS - REPETITIVE BUT PRESENT IN LOW NUMBERS) - SHARE SIMILAR SEQUENCE
NAHR - SAME ORIENTATION - DUPLICATIONS + DELETIONS
duplication of ABC region Chromosome 1
deletion of ABC region Chromosome 2.
NAHR - DIFFERENT ORIENTATION of the sequence LOW sequence NAREPEATS
reversed orientation of repeats
-can affect gene function
affects gene expression -phenotypic changes can result
NAHR - LOW COPY NUMBER REPEATS
syndrome? recurrent - happens repeatedly
NAHR - Recombination - fusion of non homologous chromosomes WITH (at) homologous sequences - unequal cross - over - duplications / deletions of regions of dna
Prader Willi syndrome - Angelman syndrome
-Duplications / deletions of low copy number repeats due to NAHR
4mB region on 15q11-q13
Phenotype depends on origin of the parent
-In the sperm or the egg?
Maternal = Prader willi - intellectual disabilities, developmental delay
Paternal = Angelman syndrome
non recurrent structural variants
recurrent = NAHR (prader willi / angel man)
non recurrent
Double strand break misrepair
DOUBLE STRAND BREAKS caused by ionising radiation / recombination
-most challenging to repair
repair of DSB
-recombination - like process
Homology directed repair e.g. CRISPR
ANOTHER WAY NHEJ - MORE ERROR PRONE
Non homologous end joining
-Broken ends are stripped back and polished - BROKEN ENDS ARE PROCESSED - (FRAYED ends must be polished)
-single strand overhangs removed
HALLMARK = presence of small deletions AT THE BREAKPOINT - (sometimes insertions)
(DSB FIXED BY NHEJ - CAUSE OF STRUCTURAL VARIATIANTS) NON RECURRENT (recurrent = NAHR insertions / deletions)
small scale deletions / insertions - Small scale variation / mutation
NAHR (recurrent) - larger scale variation than NHEJ (non-recurrent)
how else can double stranded breaks occur
-Replication fork encounters single strand break / dna lesion
-Must repair the break to PREVENT loss of genetic information
-Replication fork stalled by single strand break - leading STRAND IS STALLED
-Lagging strand continues - FORMS single DOUBLE STRAND BREAK - when replication fork collapses
-NHEJ of a double stranded broken end fused to the single DOUBLE strand break
NHEJ - is ERROR PRONE
can join to the wrong chromosome - chromosomal translocation
joined in the wrong orientation - inversions of genetic data
insertions/ deletions at the repair site
insertion,deletion,translocation,inversion (wrong orientation)
translocation - Burkitt’s Lymphoma
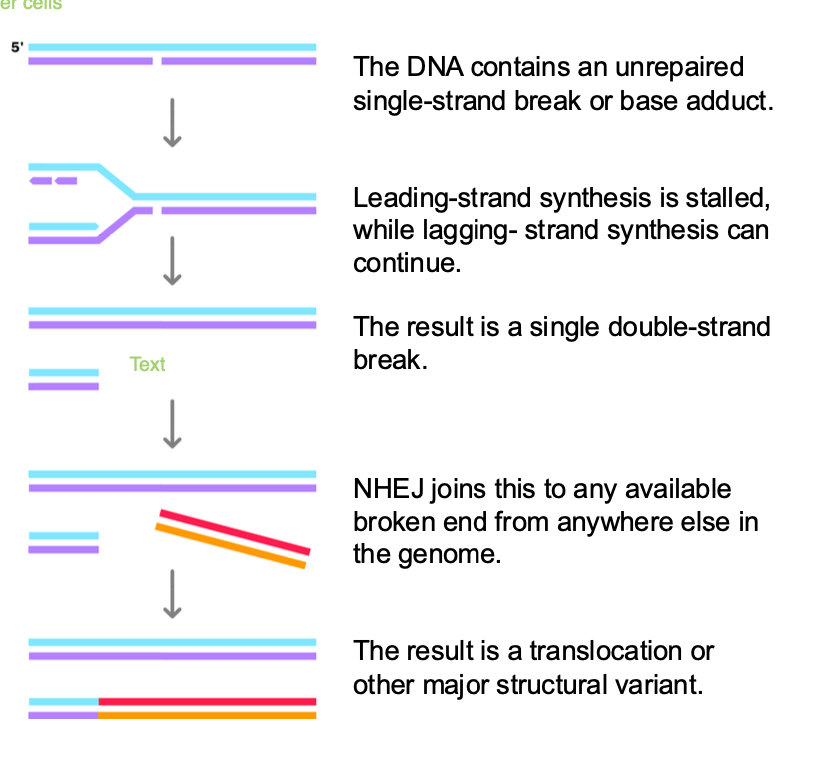
what if there is no DOUBLE strand available? for NHEJ?
-No second dsb available? - micro homology - mediated break - induced replication
3’ end of broken strand may INVADE any sequence where there is MICRO HOMOLOGY - 2-6 MATCHING BASE PAIRS
OFTEN invades DNA sequence at another replication fork with a single strand
Replication starts using this sequence
Replication process is stabilised after several cycles
-rearranged chromosome with duplications, deletions and insertions of different segments from various regions.
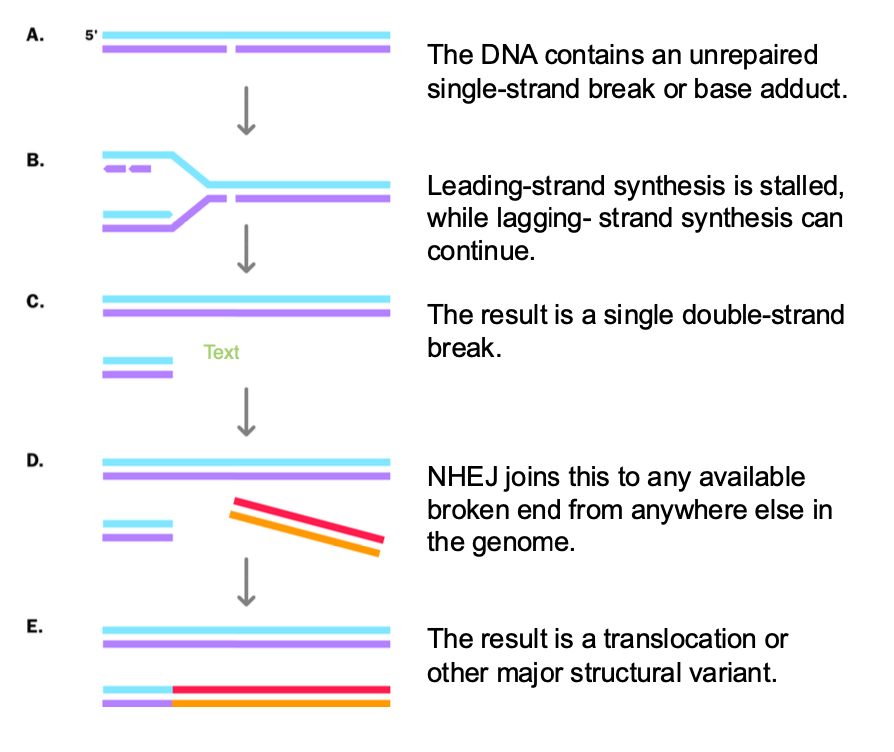
recurrent vs non recurrent
recurrent - NAHR
-ANGELMAN + PRADER WILI
NONRECURRENT - NHEJ (dsb) + Micro homology-mediated break induced replication (no 2nd dsb)
Nonreciprocal
-Only the sequence is copied from template to patch up dsb on the affected dna
Gene conversion can occur - template strand unaffected - no genetic transfer / reciprocal transfer)
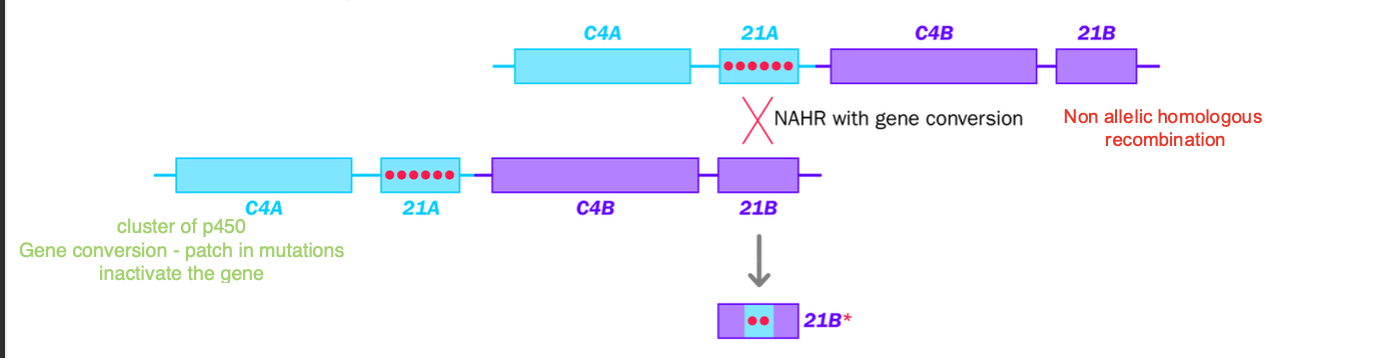
transfer is uneven - all to the broken none to the template
21B inactivated by 21A copied sequence - adrenal hyperplasia
CAH - congenital adrenal hyperplasia
Non-recurrent structural variants
Strand invasion - leading to gene conversion
Double strand break initiates normal recombination
gene conversion = non reciprocal genetic exchange
sequence from one chromosome is copied ONTO THE OTHER WITHOUT EQUAL EXCHANGE
How does it happen?
DSB occurs
-Microhomology induced recombination - 3’ end of broken dna chromosome invades a sequence of DNA with 2-6 matching base pairs
INSTEAD OF CROSS OVER RECOMBINATION
The short sequence is copied from one chromosome to another
No reciprocal transfer occurs
Original template remains unchanged
-(Invaded chromosome is unchanged, sequence is just copied
-Broken chromosome is patched with the copied sequence
21A pseudo gene into 21B
-21A pseudo gene sequence copied onto 21B
-Inactivating mutations
Makes 21B gene nonfunctional - CODING sequence is interrupted
21 hydroxylase enzyme is IMPAIRED
defective cortisol production
Excessive androgen production - adrenal hyperplasia
Conversion of the 21B gene - to inactive form
summary of lecture 3
unequal cross over in NAHR - micro deletions / duplications
DONT AFFECT THE CARRIER
-BALANCED
-Balanced abnormalities
problems during MEIOSIS
Monosomy / trisomy - aneuploidy - improper segregation
e.g. Robertsonian translocation - fine mitosis but not meiosis - trivalent structure
Segregation is improper
monosomy / trisomy due to aneuploidy - nondisjunction / anaphase lag - merotellic attachements
-Miscarriage / congenital syndrome e.g. downsyndrome

How do we find specific DNA sequences in complex mixtures?
DNA - one strand can be used to find the complementary strand
-DNA have such distinct structures - can easily be observed in complex mixtures
dna probes- detect dna
rna probes - study mrna
Nucleic Acid hybridisation
-Well characterised nucleic acid - DNA / oligonucleotide probes used to identify related sequences / complementary sequences in a complex sample
Based on Annealing
2 complementary strands of DNA will bind together to form a double stranded DNA molecule
EXAMPLES
FISH - fluorescence insitu hybridisation
fish - chromosomal abnormalities
FISH - fluorescent dna probes bind to complementary sequences - gel electrophoresis sep fragments - hybridise with dna probe + detect
-Probes detect specific chromosome - 1 signal = monosomy / 3 = trisomy - abnormalities of chromosomes
PCR primer annealing
southern blotting + northern blotting - dna digestion with restriction enzymes
Colony hybridisation
Comparative genome hybridization -test DNA + template DNA - fluorescent labelling check for deletions + duplications in the genome
RED / Green sequence lost of gained?
Denaturing and annealing dna
How is dna denatured
gentle?
Annealing?
Denaturing - double strand breaking of h.b
-Break up the hydrogen bonds in a duplex nucleic acid (double stranded)
by HEATING / exposure to highly polar chemicals e.g. urea + formamide
Separated strands can re-anneal to form the original double-stranded dna
Instead of boiling structure to denature - loss of genetic information
Instead use urea / formamide to chemically breakdown the bonds
-can use a physiologically gentle temperature - no damage of genetic information
What affects denaturing and annealing
-The extent of hydrogen bonding (denaturing is breaking up these bonds)
-Temperature - temperature can denature
-Chemical environment - highly polar can denature
Melting temperature 50% of dna dissociates into single strands due to thermal denaturation
Tm = 4°C x (number of G’s and C’s in the primer) +2°C x (number of A’s and t’S IN THE primer)
G and C have MORE HYDROGEN BONDS) G = greater
g and c = 3 hydrogen bonds
a and t = 2 hydrogen bonds
-More hydrogen bonds = higher melting temperature
Salt NACL -stabilises the hydrogen bonds - harder to denature
Strongly polar chemicals e.g. urea + formaldehyde DISRUPT the hydrogen bonds
19mer oligo with 10 gc base pairs
4×10 + 2×9 = 58 degrees
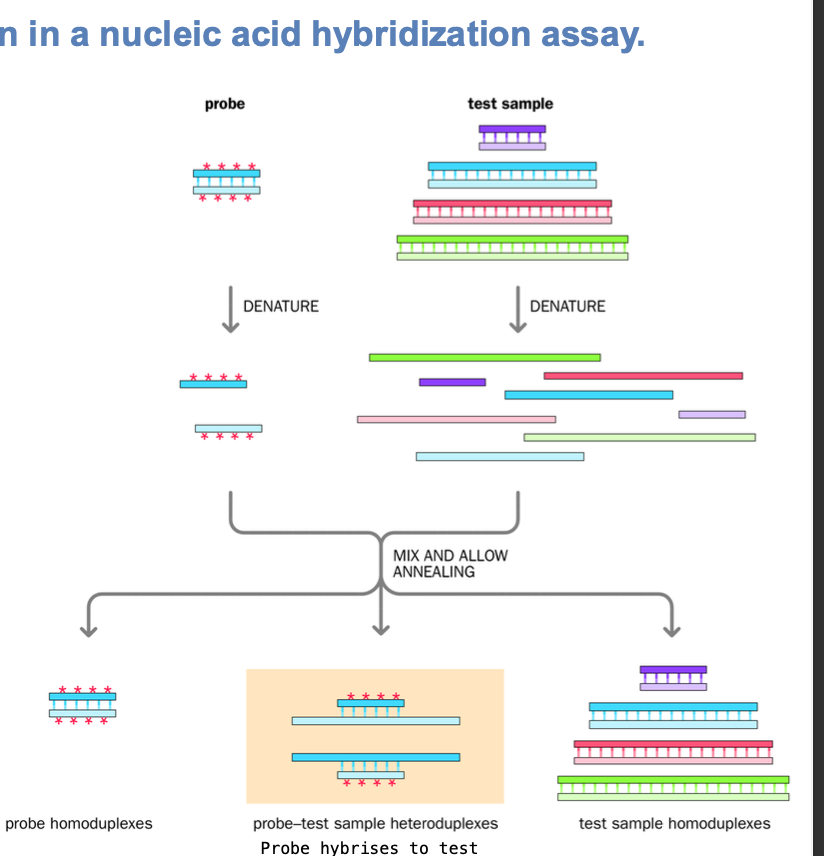
Formation of artificial duplexes + natural homoduplexes
2 DNA SOURCES - duplex = double strands
Denatured
single strands of dna ARE MIXED - homologous sequences
Homoduplex - original strands reanneal after denaturation
heteroduplex - homologous sequences on different strands re-anneal after denaturation
DNA single strands ARE MIXED UP - partially complementary sequences
heteroduplex formation in Nucleic acid hybridization assay
-DNA probe chosen - single defined nucleic acid labelled
All double strands are DENATURED
Single stranded probe nucleic acids are MIXED with single - stranded test sample nucleic acids
Strands with complementary sequences anneal
Reannealing of denatured strands - homoduplex formed
template homoduplex
probe homoduplex
ALSO
Annealing of partially complementary probe and TEST-SAMPLE SEQUENCES - HETERODUPLEX -within complex nucleic acid population
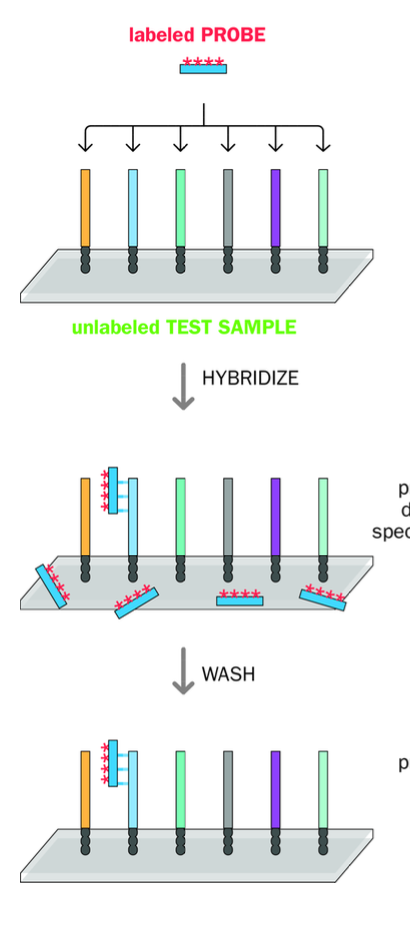
What do most hybridisation assays use?
Denatured dna samples on SOLID SUPPORTS - unlabelled
Denatured + exposed to aqueous solution of denatured labelled nucleic acid probe
Partially complementary sequences of labelled dna probe will bind to sample dna sequences
Other labelled sequences not bound - bound at incorrect locations
-CAN BE WASHED OFF
-Bound nucleic acids can be visualised in this way
Southern Blot hybridization
Blotting - requires restriction endonuclease (enzyme)
-Digest the nucleic acid
-Fragments are applied to agarose gel + separated by size using gel electrophoresis
-Gel treated with ALKALI - DENATURE THE DNA FRAGMENTS.
Placed against a nitrocellulose / nylon membrane
mEMBRANE IS SOAKED in a solution containing a radiolabelled single stranded dna probe
-After hybridization probe is washed - remove excess probe
-Membrane placed against x-ray film
Position of the labelled probe will causes a LATENT IMAGE - hybridization band
Exon sequence capture
magnet?
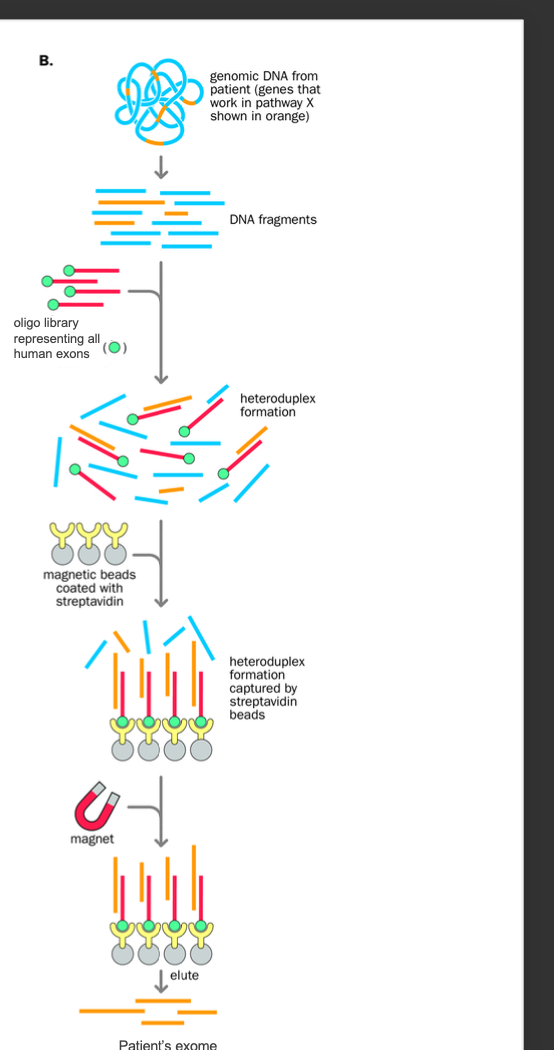
Exome
Exome = the expressed genome
BIOTINYLATED PROBE + STREPTAVIDIN COATED MAGNET
Genomic dna from patient is fragmented
Biotinylated fragment complementary to sequence of interest is prepared (biotin helps with selective purification later)
Both the fragmented dna + the biotinylated probe are denatured
Reannealing occurs - heteroduplexes form between biotinylated probe sequence + complementary sequence from the patient
Streptavidin-coated magnetic beads added
-heteroduplexes with the biotin group are bound to the beads and selectively removed using a magnet (streptavidin-coated magnet beads)
Elution of the bound patients dna to the probe
-By heating the sample - disrupts the biotin streptavidin-coated beads binding
Denature biotin dna probe+ patient dna
annealing
annealed heteroduplex retrieved by streptavidin (coated) magnetic beads
magnet removed by heating (biotinylated probe ALSO REMOVED) patients dna that has the sequence of interest remains
Random primer labelling of Nucleic acid
template dna to be used as the probe is denatured
Single strand of DNA
random primers are added - bind randomly to complementary sequences on the dna template
-dNTP mix is added AND A LABELLED NUCLEOTIDE - usally radioactively labelled
-Nucleotide is entered in region complementary to template
DNA polymerase extension
-Extends the primers
incorporates dNTPS and the labelled nucleotide
-NOW have radioactive DNA probe.
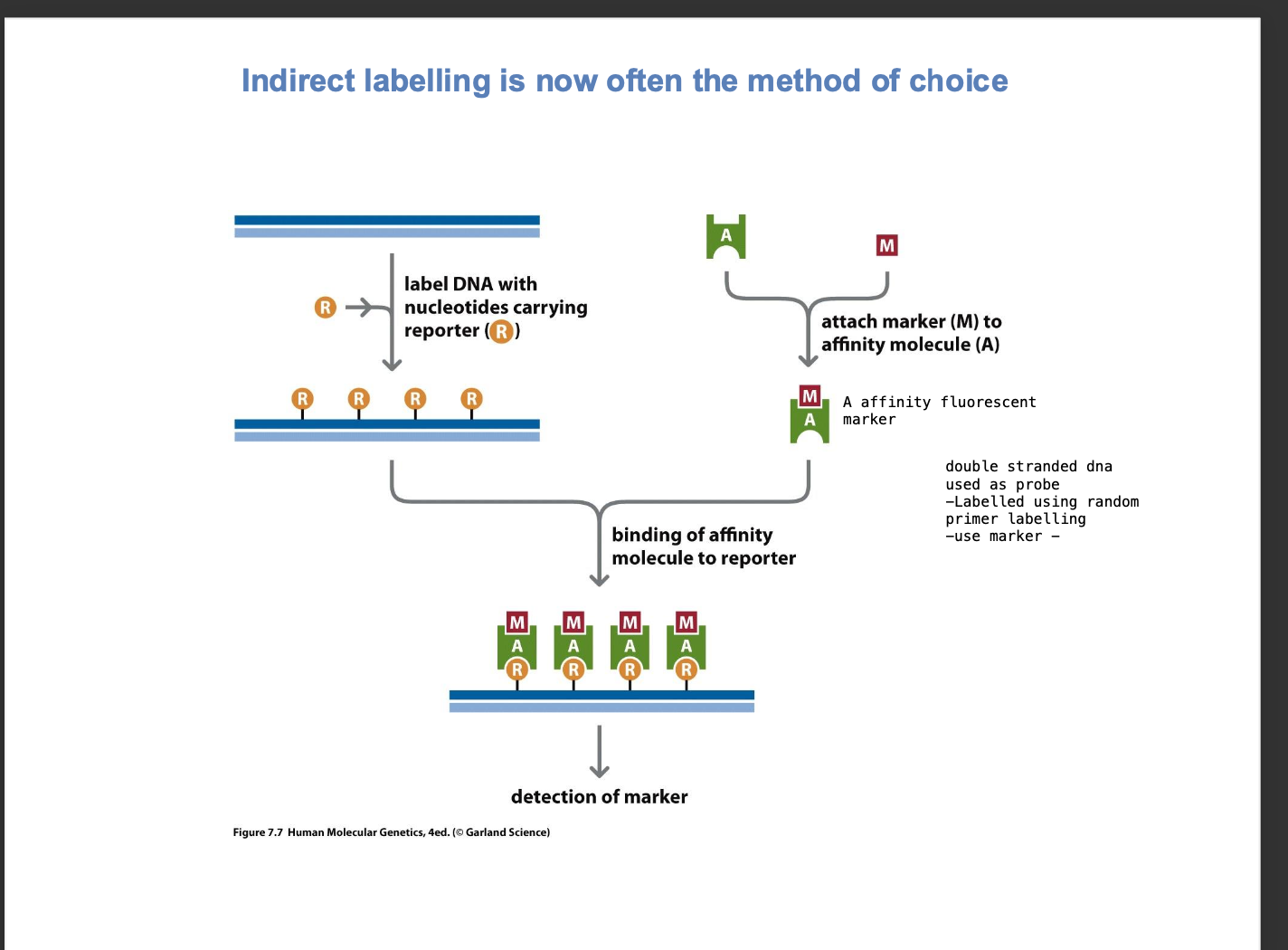
indirect labelling - method of choice
-Dna template of interest with nucleotides carrying biotin - DNA LABELLED WITH BIOTIN
-This probe is allowed to denature + bind to the sample complementary dna - sequence forming a HETERODUPLEX
-After hybridization
streptavidin coated beads - affinity molecule binds to biotin
To detect the probe - visualize the hybridized dna probe - USING REPORTERS
REPORTER = BIOTIN
AFFINITY MOLECULE = STREPTAVIDIN COATED BEADS
highly specific
DIFFERENCE TO GENOME CAPTURE
We want to visualize a genomic region
whereas here
we want to visualize the dna hybridised probe using a reporter
USES - BLOTTING LIKES PROBES
Northern blotting -DENATURE + ELECTROPHORESIS
DNA is denatured +separated by electrophoresis
DNA is hybridised with biotinylated probe
Fluorescent streptavidin beads used to visualise the probe
dna can be directly / indirectly labelled with fluorescent tags
High resolution hybridisation experiments
How can this be seen?
Indirect - reporter + affinity molecule
Direct = fluorescein / rhodamine - DNA LABELLED WITH NUCLEOTIDE
Attached to the nucleotides bound to the dna template
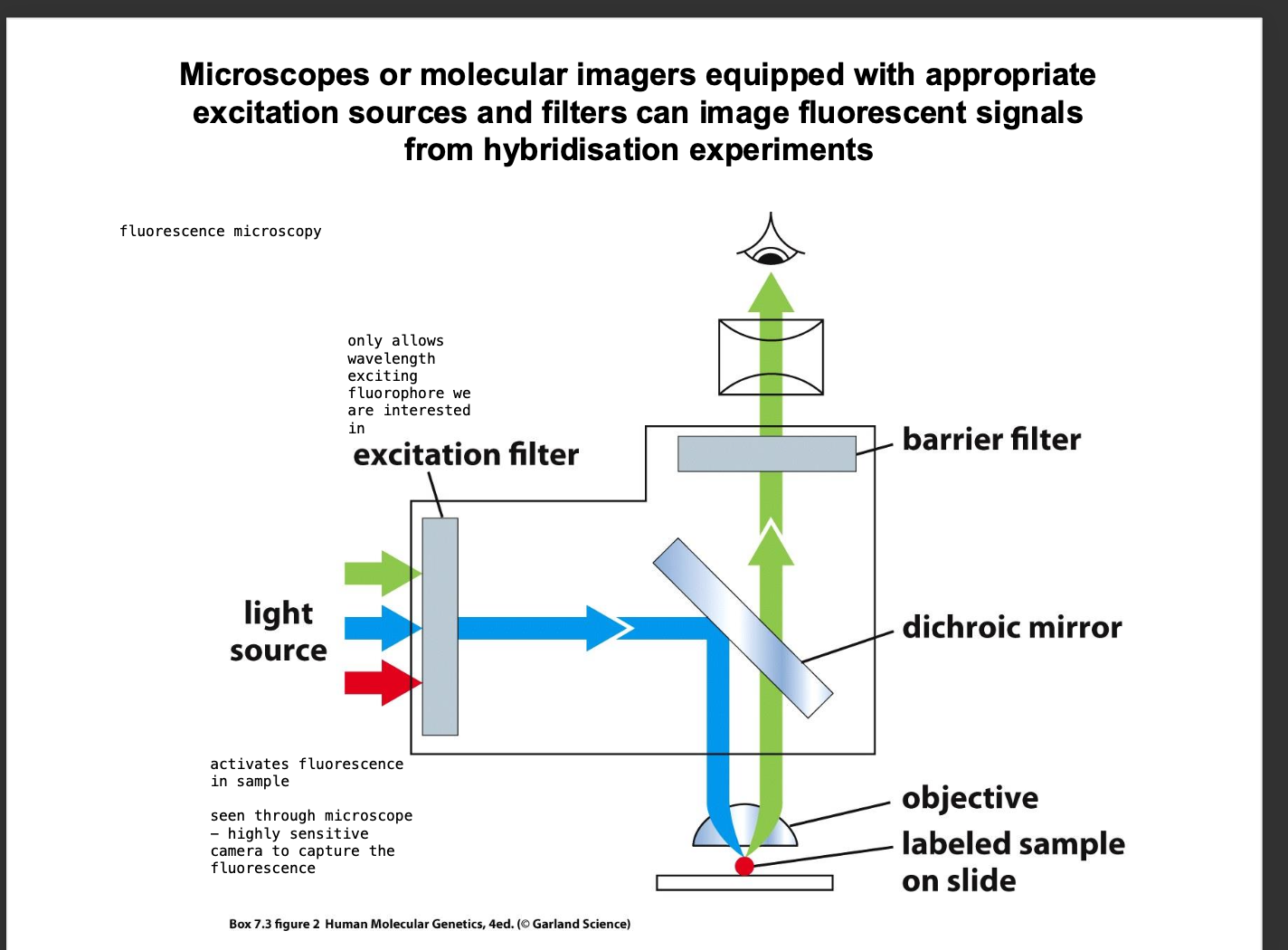
Fluorescence microscopy
-Excitation filter excites the fluorescent labelled probe
Imaging of fluorescent signals
-Equipment with APPROPRIATE EXCITATION SOURCE
-Excitation filter - wavelength of the fluorophore of interest is chosen
mages the hybridisation experiment
fluorescence microscopy
FISH
Fluorescence insitu hybridisation
-High resolution imaging microscopy
-dna probes with fluorescent marker - hybridised to sequence of interest
visualisation using fluorescence microscopy
different colour probes used for different sequences
re-annealing of single stranded dna is also dependent on?
Concentration
fixed temperature of 65 degrees celciuus
salt concentration of 0.3M NACL
High concentration of DNA = faster kinetics of re-association more complementary strands available for pairing
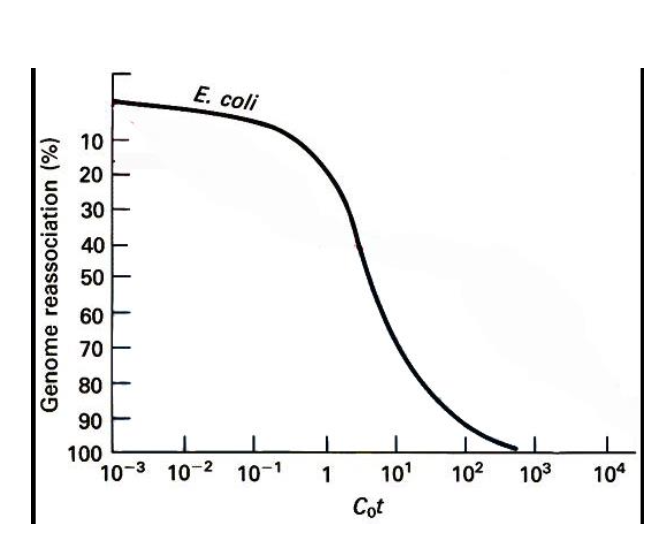
Cot curve - concentration x time
Types of sequences
Unique sequences - appear ONCE IN THE GENOME
-Reassociate quickly - do not have as many strands to form repetitive regions
Repetitive sequences - Reassociate slowly
-Many copies of the same sequence are available for reassociation
They appear LATER IN THE COT CURVE
SLOWER reassociation rate
CoT curves
-What is measured
-What is the rate limiting step?
DENATURE + REANNEAL
-Shearing dna to 1.000 bp length
denaturing the dna with heat
lowering the temp to allow re annealing
Measure the % still single stranded at various time points
Rate limiting step?
-Collision of 2 complementary molecules - (formation of the double strand)
2nd order kinetics - rate of the association = initial concentration x time
reannealing - producs a Sigmodial curve - S SHAPED CURVE
-Characteristics of CoT analysis
S shaped curve
-Early on - conc of single stranded DNA is high
Later on - conc of single stranded dna is much lower
CoT ½ value - the point where ½ of the DNA is still single stranded
Unique sequences - CoT 1/2 value is much lower - faster reannealing
Repetitive sequences CoT ½ sequence is much higher - slower reannealing
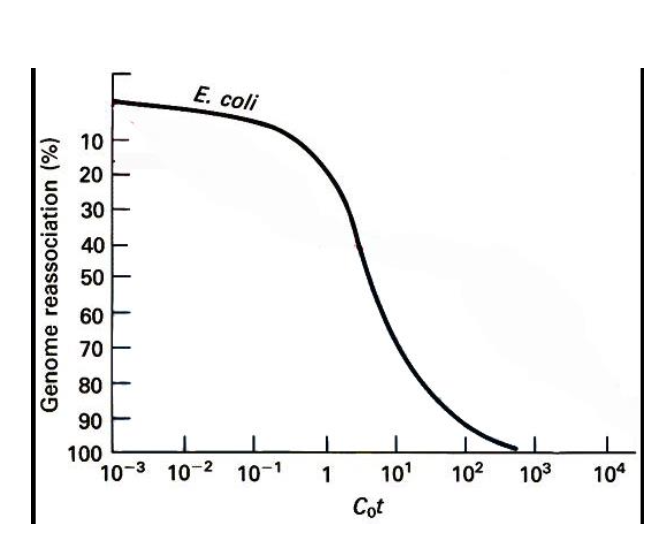
CoT curve in the human genome
Complex
Not simple sigmoid
3 SECTIONS
Highly repeated DNA - about 50,000 copies - high CoT
Moderately repeated DNA 500 copies - lower CoT
Unique dna - 10 copies - Lowest CoT
-Each component has its own CoT ½ value - represents a characteristic portion of the genome
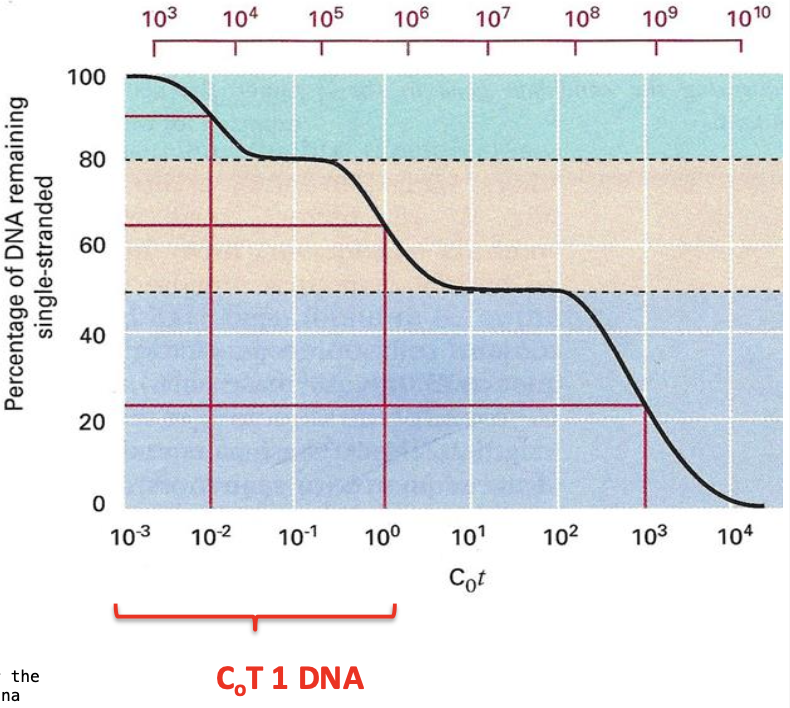
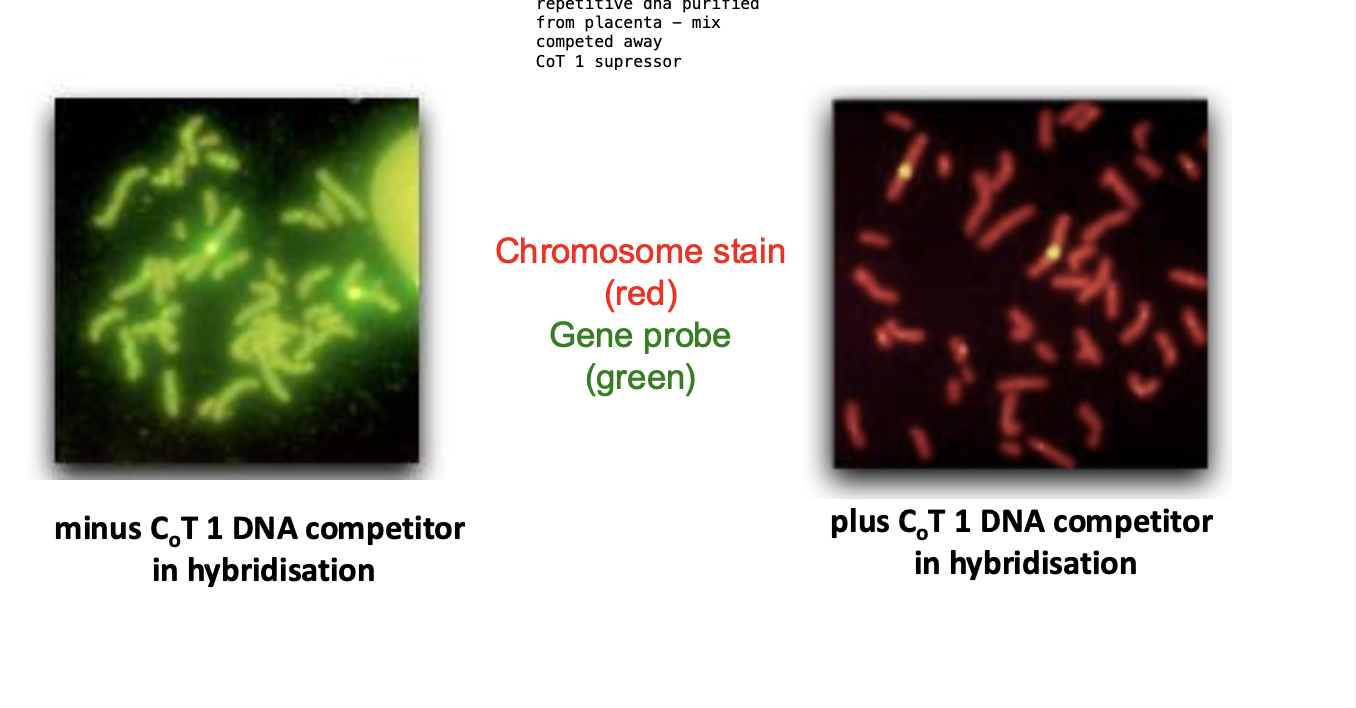
Highly repetitive elements of DNA are known as?
Use in Fish?
Highly repetitive DNA = CoT 1 DNA SUPPRESSOR
IN FISH
CoT 1 DNA
Prevents the probe from binding to repetitive DNA sequences
competes with the probe for binding sites
Improves signal-noise reatio
REDUCES THE BACKGROUND SIGNAL
IN FISH
Cot 1 DNA - added to solution before the probe
EASIER TO DETECT TARGET SEQUENCES
Microarray hybridisation
MICRO array
-grid of Immobilised probes in an array
High- throughput technique
-SOLUTION WITH SINGLE STRANDED DNA SEQUENCES
IMMOBILISED PROBES ASSAY IN GRID FORMAT
simultaneous analysis of thousands of sequences
Hybridising ARRAY of test sample nucleic acids to ARRAY OF MMOBILISED PROBES arranged in a grid
Each probe is specific to a sequence
Aqueous solution containing a Heterogenous collection of LABELLED DNA FRAGMENTS OR RNA TRANSCRIPTS
Denatures + allowed to hybridize with the probes on the array
Some probes - will have lots of sequences complementary to the dna sequences in the solution -
-other probes - few complementary sequences in the test sample - WEAK HYBRIDIZATION SAMPLE
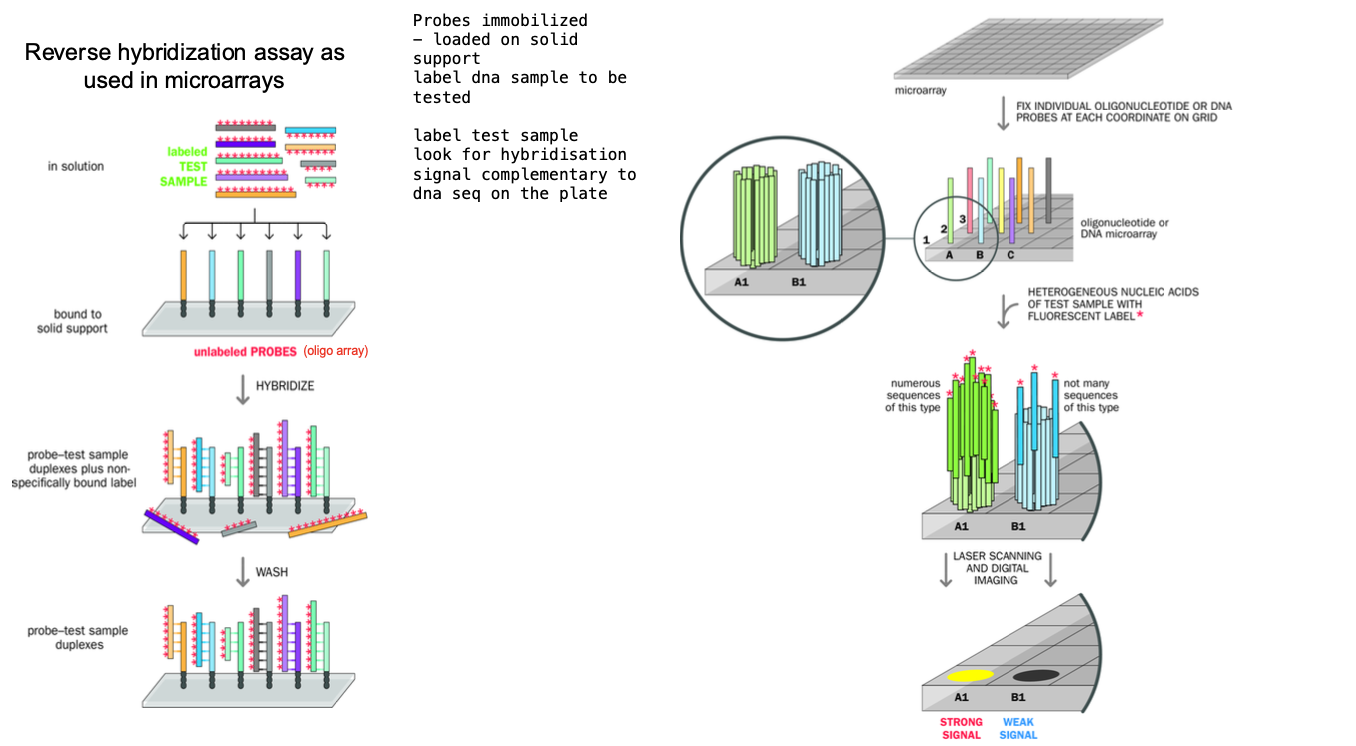
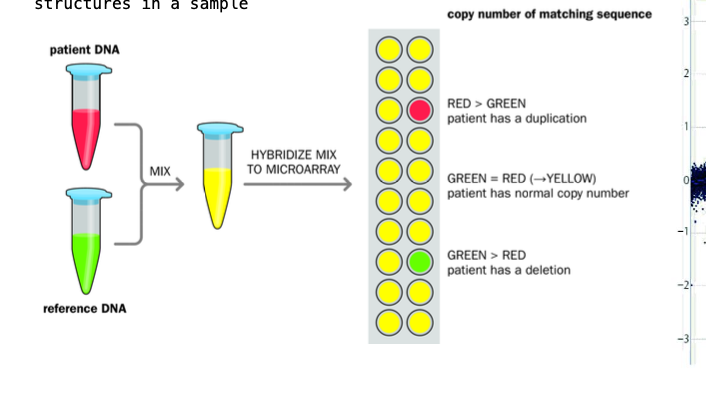
Array comparative genomic hybridisation - ACGH
Reference DNA sample + test patient - labelled with different fluorophores
Fragmented using restriction endonucleases
-Denatured - made single stranded
Mixed in equal genomic amount + ALLOWED TO HYBRIDISE to the microarray containing KNOWN GENOMIC SEQUENCES (probe spot)
reference dna and the patient dna COMPETE FOR each probe spot
Reference = green, tested patients dna = red
Colour of the cell of the array AFTER hybridisation - measure of the relative amounts of the dna in each spot of the array
Yellow = even mix of the 2 - 2 normal copies of DNA in the sequence (green and red makes yellow)
Red = duplication - gain of genetic material e.g. 3 copies of a chromosome
green = deletion - loss of genetic material (the reference dna dominates) e.g. 1 copy of a chromosome
TESTING FOR GENETIC DISORDER, COPY NUMBER VARIATION, CANCER DIAGNOSTICS
PCR
Polymerase chain reaction
-Design of primerr forward and back - annealing of the primers
amplification of the primers
-make sure the primers dont sit in regions of highly repetitive DNA
-want to avoid repetitive elements in the genome
Taq polymerase - heat stable is used
denaturation, annealing, extension
Template dna, primers - short synthetic single strands
taq polymerase
buffer and mg
Primer design
what is important - how can we fix this?
-Want to avoid primers in regions of highly repetitive dna
dont want to amplify already highly repetitive dna
FOCUS ON THE UNIQUE DNA SEQUENCE
REPEAT MASKER
-Identifies repetitive elements + removes them or masks them
dna cloning can be done using?
Plasmid vectors
cut the plasmid vector with a restriction enzyme
replace the section in the plasmid with the human DNA to be cloned - using dna ligase to join them together
transformation - bacteria grown in a media where ONLY THE RECOMBINANT PLASMIDS CAN SURVIVE
antibiotic resistance in plasmids
amplification of dna
BACTERIA CULTURED IN LARGE QUANTITIES
BAC
Making bac
BAC LIBRARIES
bacterial artificial chromosome vectors
Vector based on the
BAC VECTORS - enormous capacity
MBO 1 - cuts up the DNA of interest into fragments
Compatible “sticky ends” with Bam H I of the plasmid vector
genome projects - cloning of large regions of dna
Large inserts possible
DNA ISOLATED
DNA fragmented using restriction enzymes - MBo 1 -
then undergo pulsed gel electrophoresis
-dna fused to plasmid with dna ligase - mbo 1 sticky end to Bam HI
blue white screening - identifies colonies with INSERTS
Antibiotic selection - only bacteria with plasmid survive
-Robotic arrays speed up the process - can represent 90% of the genome
each 4×4 box - 8 clones plated in duplicate
summary
-comparative array genomic hybridisation
micro array hybridiation (immobilised probes)
cot1 dna - supressor of background noise for (repetitive dna)
SOUTHERN BLOT - FOR DNA
Restriction - electrophoreisis -hybridisation

What do genetic maps rely on?
Principle of genetic maps - genes located closely on the same chromosome are expected to be inherited together.
RFLP
1st genetic maps - constructed using polymorphic dna markers
How is polymorphism detected now?
-Restriction fragment length polymorphism - to detect polymorphic markers
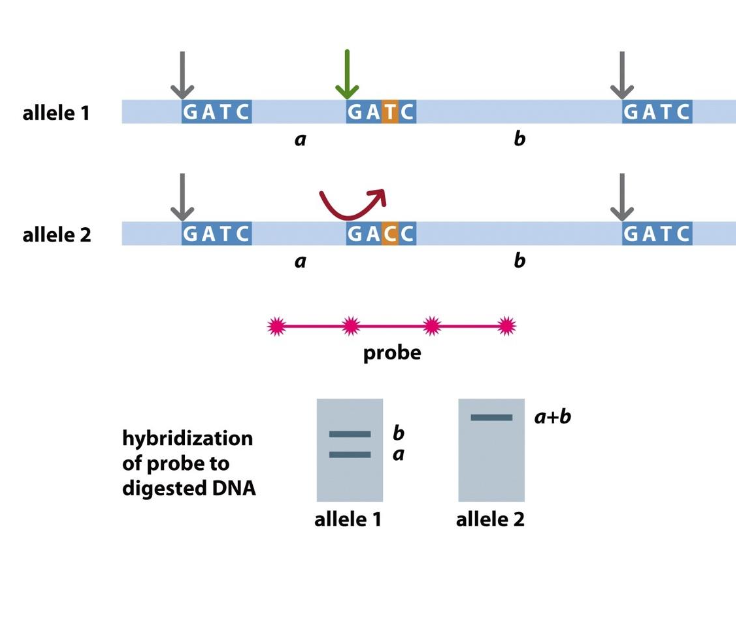
A polymorphic marker is a region of DNA that varies between individuals (e.g., a mutation changing GATC → GACC). - detected by restriction enzymes (fragments)
Detect variation
MBOL - detects normal sequence GATC
-Allele 1 - has cut at GATC - Mbol cuts here
Allele 2 - has variation - not the normal GATC sequence - Mbol can’t cut here
In the past
-difference would be highlighted using a SOUTHERN BLOT ASSAY
-Mbol digests
fragments separated by electrophoresis
dna probe used to find the desired sequences
If has the restriction enzyme sequence - 2 fragment appears (cut at this site)
if not there is no cut
Now polymorphism detected using PCR
Cut using restriction eznymes - separate the fragments by electrophoresis
393 RFLP used for genetic linkage map of the human genome
AMPLIFICATION OF POLYMORPHISMS USING PCR
DIGESTION USING RESTRICTION ENZYME mbol
Second generation human genetic map - based on microsatellite DNA
Microsatellite DNA - aka Short tandem repeats
Instability arises - DURING MISTAKES IN REPLICATION
Polyacrylamide gel electrophoresis is used - see VARIATION IN SATELLITE DNA
-High resolution for small fragments of DNA
Separation based on size
-NUMBER OF REPEATS VARIES BETWEEN INDIVIDUALS - Good for genetic finger printing
Assayed by PCR - CREATE LARGE AMPLIFICATIONS TO TEST FOR polymorphisms in the satellite dna
Polymorphism = variation that occurs in the dna sequences among individuals in a population
Amplify using PCR
Separation for analysis using PAGE
Weissenbach et al - segregation analysis of 814 polymorphic locai containing (C-A)n repeats from 8 large families
Analysis of the sequences of dna
FORENSIC dna profiling
dna testing