GIO: L5 02-10 CONTROL OF GENE EXPRESSION
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
33 Terms
WHAT CONTAIN THE SAME GENOME?
DIFFERENT CELL TYPES!
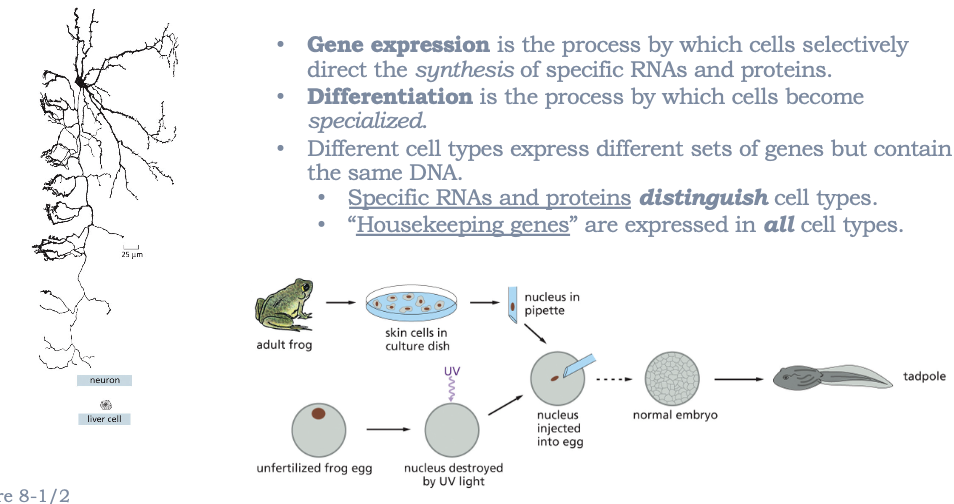
GENE EXPRESSION IS THE PROCESS BY WHICH CELLS SELECTIVELY DIRECT THE SYNTHESIS OF SPECIFIC RNAS AND PROTEINS
DIFFERENTIATION IS THE PROCESS BY WHICH CELLS BECOME SPECIALIZED
DIFFERENT CELL TYPES EXPRESS DIFFERENT SETS OF GENES BUT CONTAIN THE SAME DNA
SPECIFIC RNAS AND PROTEINS DISTINGUISH CELL TYPES
“HOUSEKEEPER GENES” ARE EXPRESSED IN ALL CELL TYPES
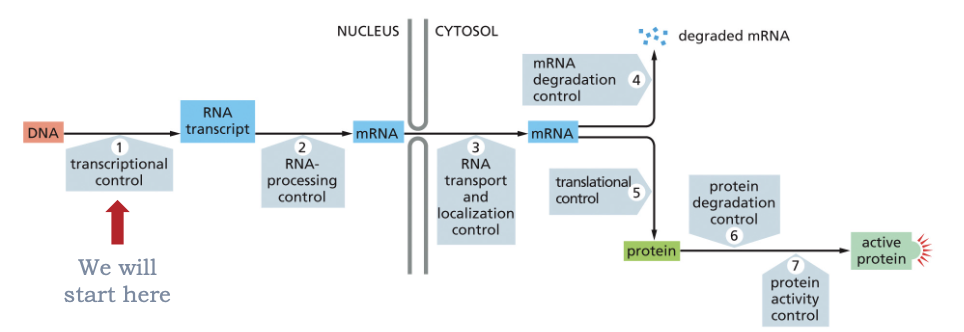
WHAT CAN BE REGULATED AT MULTIPLE STEPS?
GENE EXPRESSION!
TRANSCRIPTION IS REGULATED IN BOTH PROKARYOTES AND IN EUKARYOTES
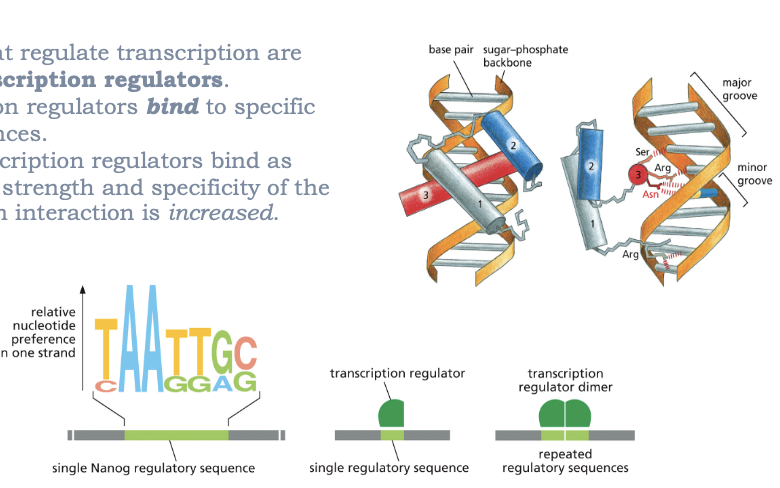
WHAT BIND TO DNA?
TRANSCRIPTION REGULATORS!
PROTEINS THAT REGULATE TRANSCRIPTION ARE CALLED TRANSCRIPTION FACTORS
TRANSCRIPION REGULATORS BIND TO SPECIFIC DNA SEQUENCES
WHEN TRANSCRIPTION REGULATORS BIND AS DIMERS, THE STRENGTH AND SPECIFICITY OF DNA-PROTEIN INTERACTION IS INCREASED
WHAT ARE ARRANGED IN OPERONS?
PROKARYOTIC GENES
THE TRP (TRYPTOPHAN) OPERON IS AN EXAMPLE OF OPERONS
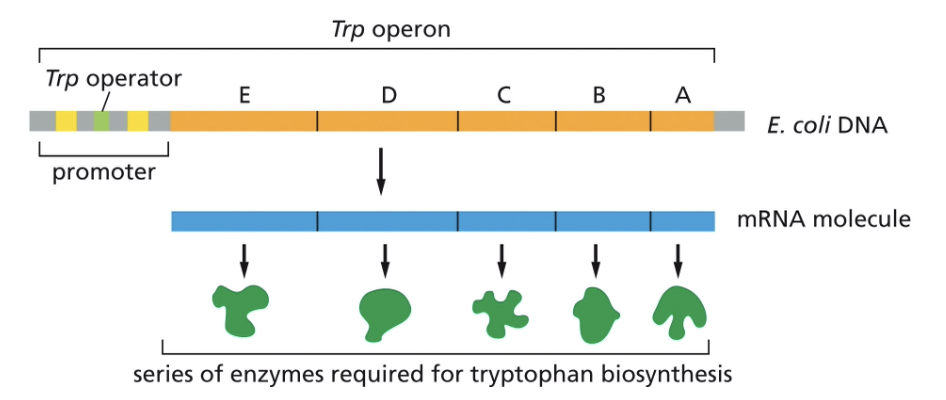
GENES CAN BE SWITCHED OFF BY…
REPRESSOR PROTEINS
THE LEVELS OF TRYPTOPHAN REGULATE THE EXPRESSION OF THE OPERON
THE TRP REPRESSOR IS A TRANSCRIPTIONAL REPRESSOR
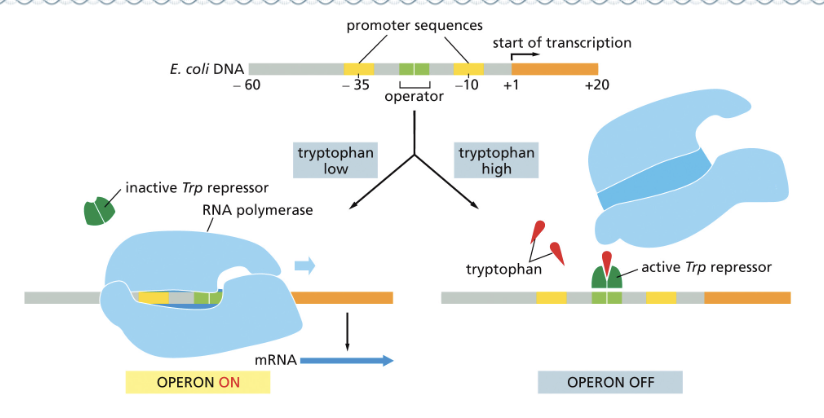
GENE EXPRESSION CAN BE SWITCHED ON BY…
ACTIVATOR PROTEINS
ACTIVATOR PROTEINS PROMOTE THE BINDING OF RNA POLYMERASE
REPRESSOR PROTEINS INTERFERE WITH THE BINDING OF RNA POLYMERASE
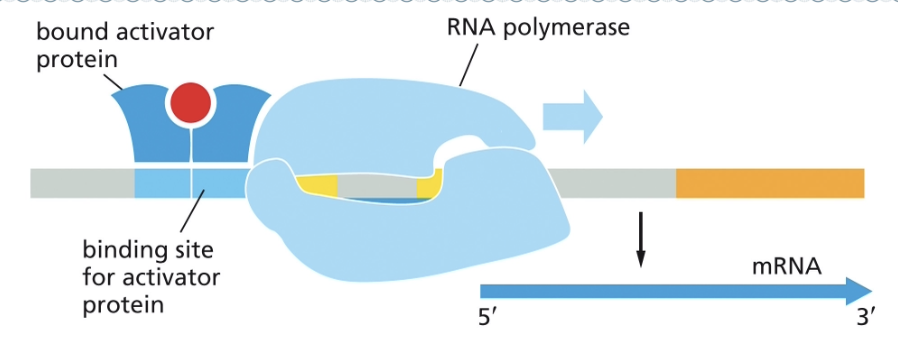
WHAT IS TRANSCRIPTIONAL REGULATION IN PROKARYOTES?
IN PROKARYOTES:
BOTH ACTIVATORS AND REPRESSORS REGULATE TRANSCRIPTION
WHAT IS TRANSCRIPTIONAL REGULATION IN EUKARYOTES?
IN EUKARYOTES
PROMOTER ACCESS IS RESTRICTED
CONDENSED CHROMATIN IS INACCESSIBLE
CHROMATIN MUST BE REMODELED
TRANSCRIPTION REGULATORS CAN RECRUIT CHROMATIN-REMODELING PROTEINS TO ACTIVATE TRANSCRIPTION
ACTIVATION CAN OCCUR AT A DISTANCE
ENHANCERS RECRUIT CHROMATIN-REMODELING ENZYMES
ENHANCERS RECRUIT TRANSCRIPTIONAL ACTIVATORS
WHAT CAN BE CONTROLLED AT A DISTANCE?
EUKARYOTIC GENE ACTIVATION!
EUKARYOTIC TRANSCRIPTIONAL REGULATORS BIND ENHANCERS (A DNA SEQUENCE)
THE MEDIATOR COMPLEX LINKS ACTIVATORS AND THE GENERAL TRANSCRIPTION MACHINERY TO PROMOTE (OR REPRESS) ASSEMBLY
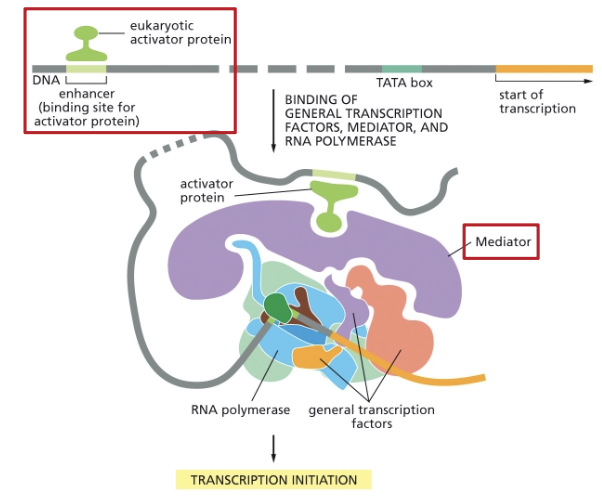
WHAT MEDIATE ENHANCER-PROMOTOR INTERACTIONS?
DNA LOOPS!
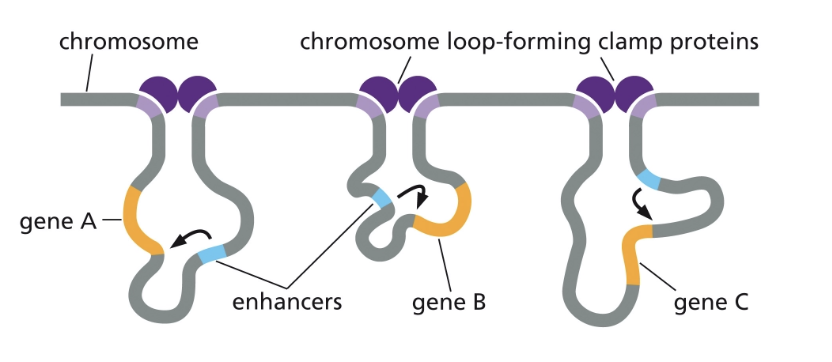
EUKARYOTIC GENE TRANSCRIPTION REGULATORS CAN…
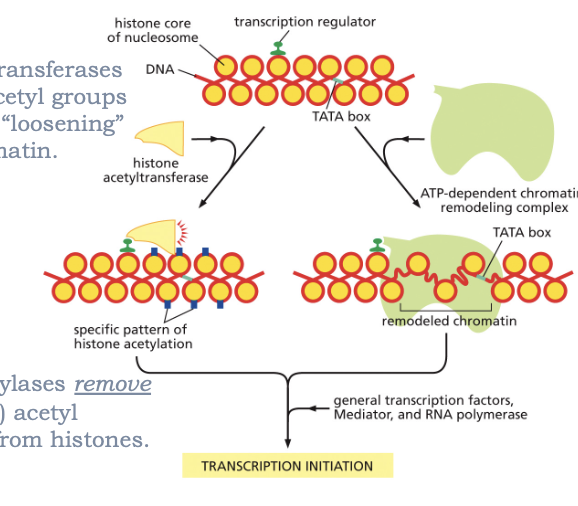
RECRUIT CHROMATIN MODIFYING PROTEINS!
HISTONE ACETYLTRANSFEREASES ADD (WRITERS) ACETYL GROUPS TO HISTONE TAILS “LOOSENING” THE CHROMATIN
HISTONE DEACETYLASES REMOVE (ERASERS) ACETYL MODIFICATIONS FROM HISTONES
ATP-DEPENDENT CHROMATIN REMODELING COMPLEXES REPOSITION DNA ON NUCLEOSOMES
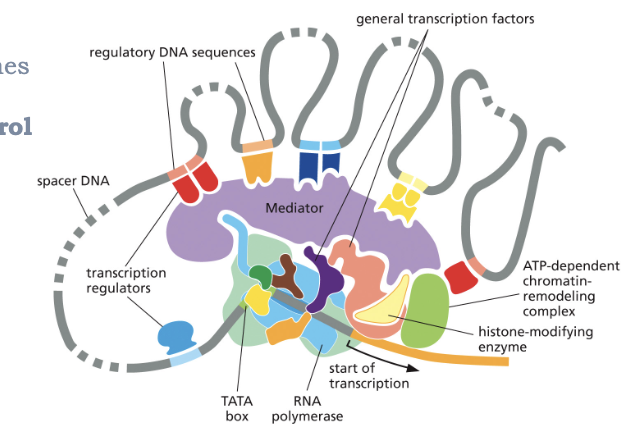
WHAT IS COMBINATORIAL CONTROL OF TRANSCRIPTION?
MANY EUKARYOTIC GENES ARE UNDER COMBINATORIAL CONTROL BY MULTIPLE TRANSCRIPTION REGULATORS
TOGETHER, THESE REGULATORS DETERMINE THE RATE OF TRANSCRIPTION
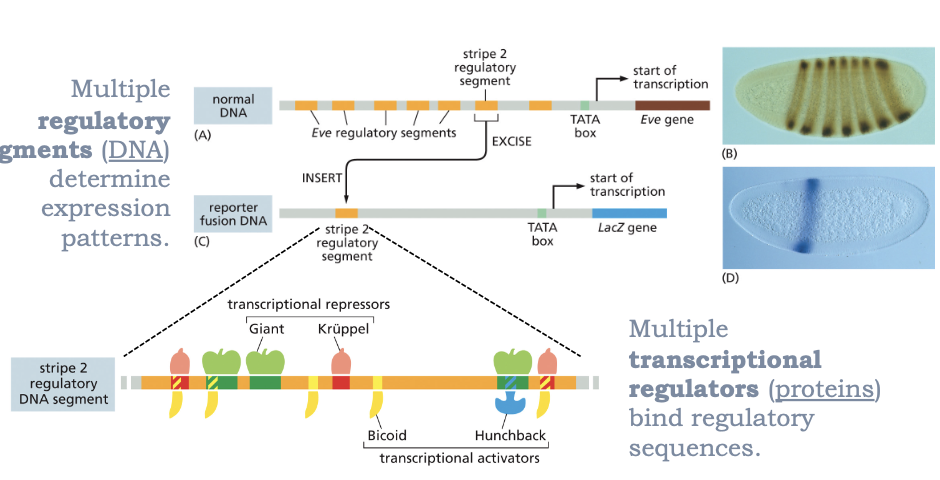
WHAT IS COMBINATORIAL CONTROL OF TRANSCRIPTION: THE EXAMPLE OF EVE?
MULTIPLE TRANSCRIPTIONAL REGULATORS (PROTEINS) BIND REGULATORY SEQUENCES
MULTIPLE REGULATORY SEGMENTS (DNA) DETERMINE EXPRESSION PATTERNS
WHAT CAN COORDINATE EXPRESSION OF MANY GENES?
A SINGLE TRANSCRIPTIONAL REGULATOR!
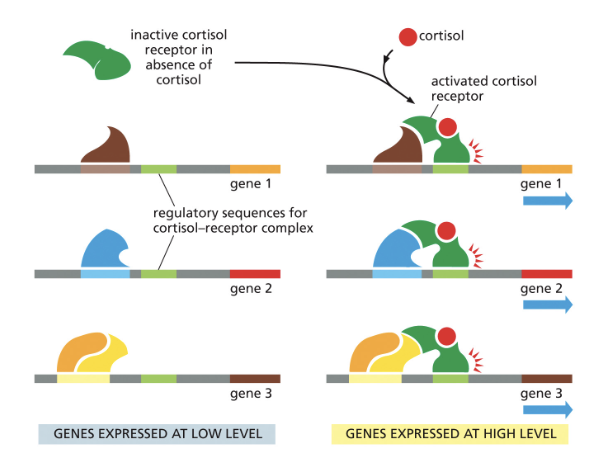
EXAMPLE: CORTISOL RECEPTOR
CORTISOL RECEPTOR US A TRANSCRIPTIONAL REGULATOR
MANY COPIES OF THE CORTISOL RECEPTOR ARE PRESENT IN A SINGLE LIVER CELL
MANY GENES CONTAIN THE SAME CORTISOL RECEPTOR REGULATORY SEQUENCE
IN THE PRESENCE OF CORTISOL, THE CORTISOL RECEPTOR BINDS TO THE DNA AND INTERACTS WITH TRANSCRIPTIONAL ACTIVATORS TO PROMOTE HIGH LEVELS OF TRANSCRIPTION
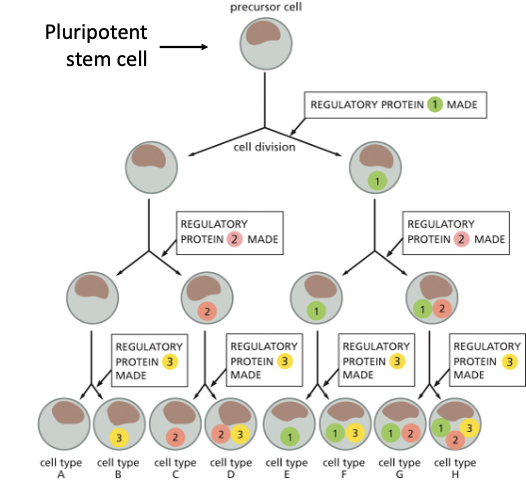
DIFFERENT COMBINATIONS OF TRANSCRIPTIONAL REGULATORS CAN…
GENERATE DIFFERENT CELL TYPES!
HYPOTHETICAL SCHEME: 7 DIFFERENT CELL TYPES ARE GENERATED USING 3 DIFFERENT TRANSCRIPTIONAL REGULATORS
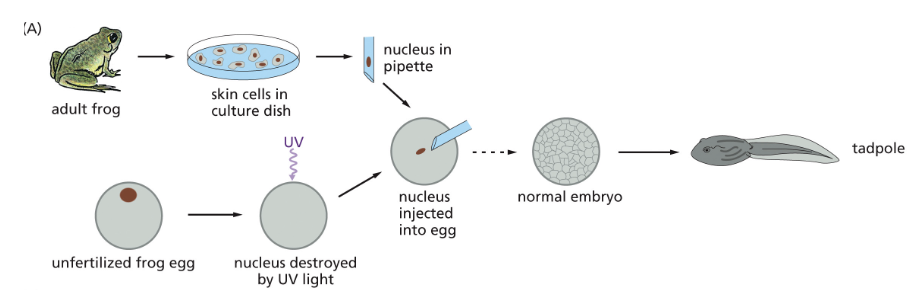
WHAT IS CELLULAR PROGRAMMING?
DIFFERENTIATED CELLS CONTAIN ALL THE GENETIC INSTRUCTIONS NEEDED TO DIRECT THE FORMATION OF A COMPLETE ORGANISM
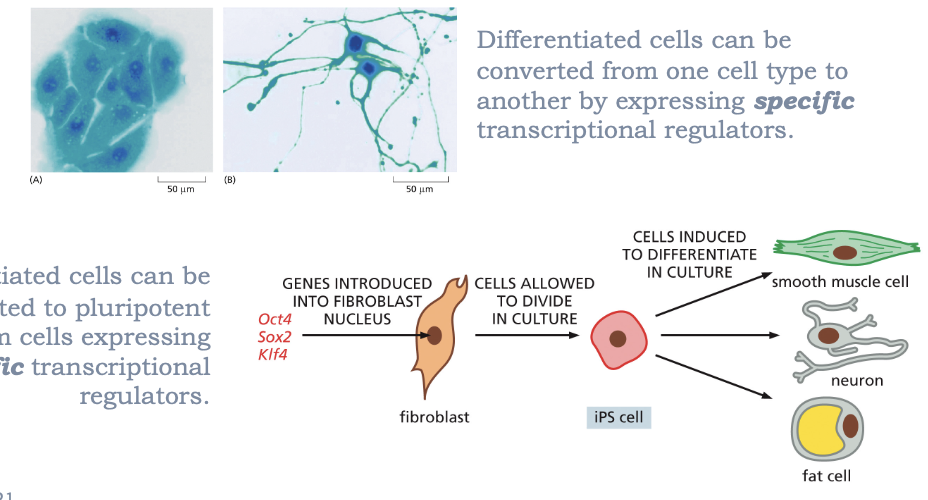
WHAT IS CELLULAR REPROGRAMMING?
DIFFERENTIATED CELLS CAN BE CONVERTED FROM ONE CELL TYPE TO ANOTHER BY EXPRESSING SPECIFIC TRANSCRIPTIONAL REGULATORS
DIFFERENTIATED CELLS CAN BE CONVERTED TO PLURIPOTENT STEM CELLS EXPRESSING SPECIFIC TRANSCRIPTIONAL FACTORS
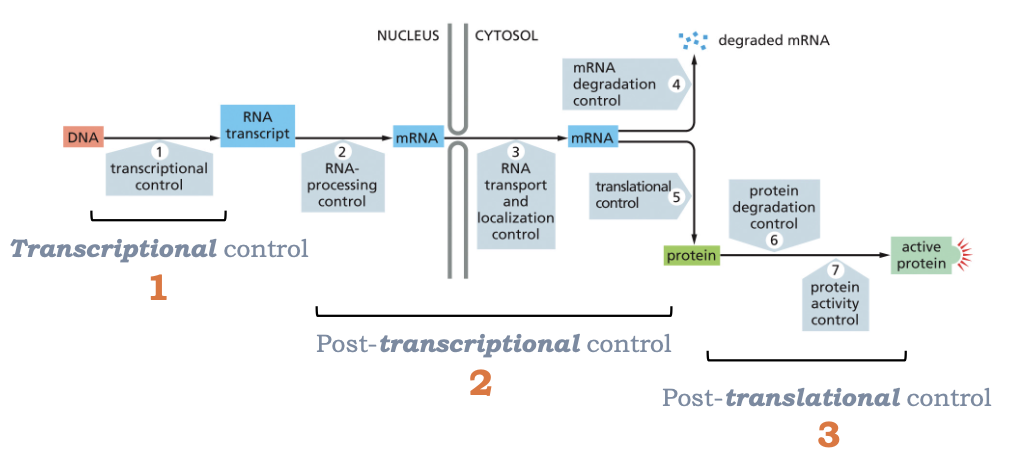
WHAT ARE THE MULTIPLE STEPS THAT GENE EXPRESSION CAN BE REGULATED AT?
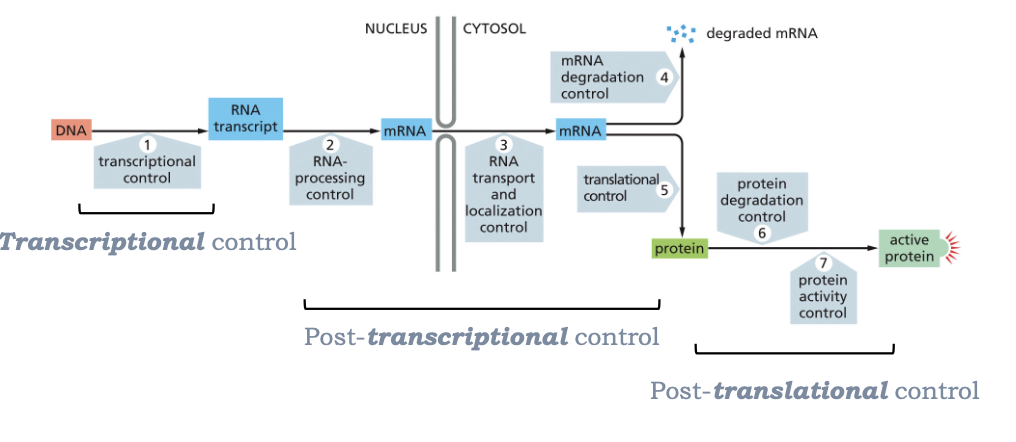
WHAT IS THE FUNCTION OF MESSENGER RNAS (MRNAS)?
CODE FOR PROTEINS
WHAT IS THE FUNCTION OF RIBOSOMAL RNAS (RRNAS)?
FORM THE CORE OF THE RIBOSOME’S STRUCTURE AND CATALYZE PROTEIN SYNTHESIS
WHAT IS THE FUNCTION OF MICRORNAS (MIRNAS)?
REGULATE GENE EXPRESSION
WHAT IS THE FUNCTION OF TRANSFER RNAS?
SERVE AS ADAPTORS BETWEEN MRNA AND AMINO ACIDS DURING PROTEIN SYNTHESIS
WHAT IS THE FUNCTION OF SMALL INTERFERING RNAS (SIRNAS)?
PROVIDE PROTECTION FROM VIRUSES AND PROLIFERATING TRANSPOSABLE ELEMENTS
WHAT IS THE FUNCTION OF LONG NONCODING RNAS (LNCRNAS)?
ACT AS SCAFFOLDS AND SERVE OTHER DIVERSE FUNCTIONS, MANY OF WHICH ARE STILL BEING DISCOVERED
WHAT IS THE FUNCTION OF OTHER NONCODING RNAS?
USED IN RNA SPLICING, GENE REGULATION, TELOMERE MAINTENANCE, AND MANY OTHER PROCESSES
WHAT TARGET RNAS FOR DEGRADATION?
MICRORNAS (MIRNAS) CONTROL GENE EXPRESSION THROUGH BASE-PAIRING INTERACTIONS
MATURE MIRNAS ARE -22 NTS IN LENGTH
SINGLE-STRANDED MIRNAS FORM A COMPLEX WITH PROTEINS TO FORM A RNA-INDUCED SILENCING COMPLEX (RISC)
RISC BINDS TO COMPLEMENTARY RNA SEQUENCES
RESULT: DEGRADATION OF THE TARGET MRNA
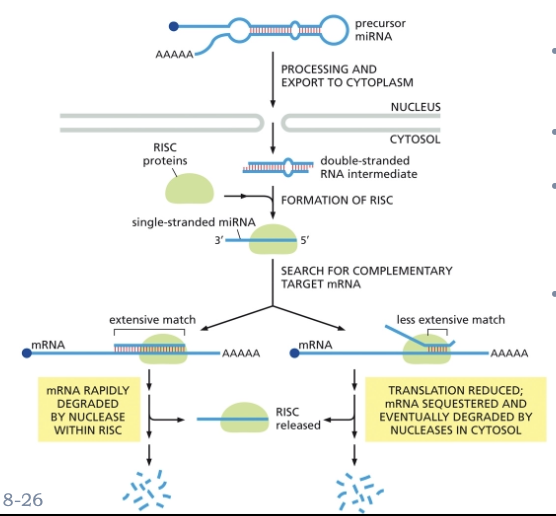
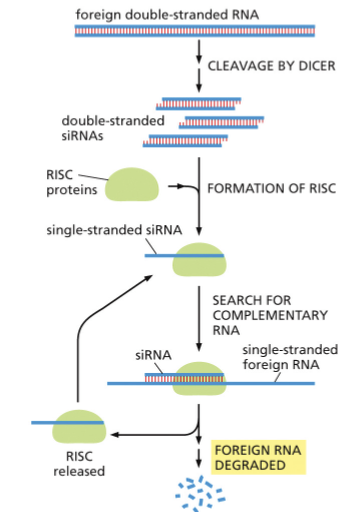
WHAT IS GENE SILENCING BY RNA INTERFERENCE?
SMALL INTERFERING RNAS (SIRNAS) ARE PRODUCED FROM DOUBLE-STRANDED FOREIGN RNAS
DICER IS AN ENDONUCLEASE THAT CLEAVES FOREIGN DOUBLE-STRANDED RNAS
SINGLE-STRANDED SIRNAS FORM A COMPLEX WITH PROTEINS TO FORM AN RNA-INDUCED SILENCING COMPLEX (RISC)
RISC BINDS TO COMPLEMENTARY RNA SEQUENCES
RESULT: DEGRADATION OF THE TARGET MRNA
WHO DISCOVERED RNA INTERFERENCE?

WHO DISCOVERED MICRORNAS?

WHAT ARE CAENORHABDITIS ELEGANS?
WHAT IS POST-TRANSLATIONAL MODIFICATION OF PROTEINS?
POST-TRANSLATIONAL MODIFICATIONS INCLUDE:
1) METHYLATION
2) ACETYLATION
3) PHOSPHORYLATION
4)UBIQUITINATION
5)GLYCOSYLATION
MODIFICATIONS CAN AFFECT BINDING TO CO-FACTORS, STABILITY, AND SUBCELLULAR LOCALIZATION
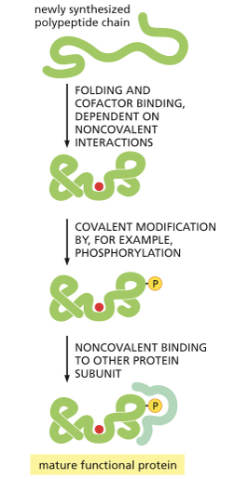
WHAT IS REGULATION OF PROTEIN DEGRADATION?
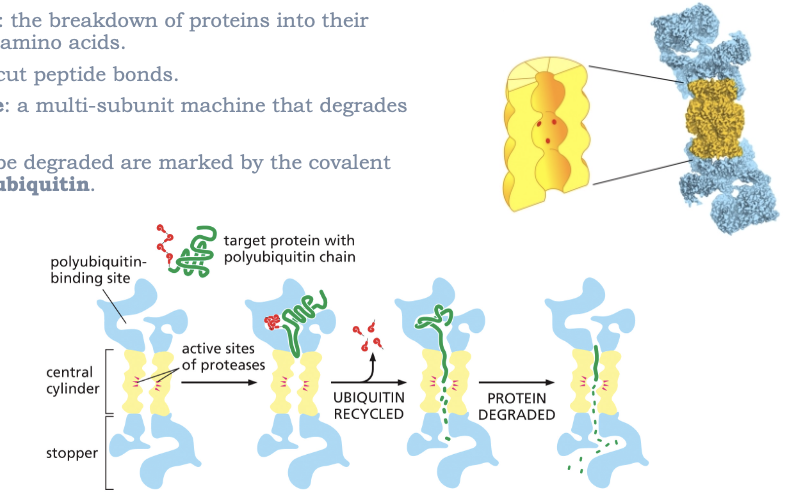
PROTEOLYSIS: THE BREAKDOWN OF PROTEINS INTO THEIR CONSTITUENT AMINO ACIDS
PROTEASES: CUT PEPTIDE BONDS
PROTEASOME: A MULTI-SUBUNIT MACHINE THAT DEGRADES PROTEINS
PROTEINS TO BE DEGRADED ARE MARKED BY THE COVALENT ADDITION OF UBIQUITIN
WHAT ARE THE 3 STEPS THAT GENE EXPRESSION CAN BE REGULATED?