A Level CIE Biology: 17 Selection & Evolution
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
99 Terms
phenotype
observable characteristics of an organism
phenotypic variation
difference in phenotypes between organisms of the same species
how can phenotypic variation be explained
genetic factors e.g. 4 diff bloo groups bc 3 possible alleles
environmental factors e.g. diff heights of plant in diff conditions
combination of both e.g. recessive allele for sickle cell has high freq where malaria is prevalent bc resistance if heterozygous
complete phenotype of organism equation
genotype + environment
genetic variation
organisms of same species have v similar genotypes but 2 individuals (even twins) will have differences between their DNA base sequences
considering size of genomes, these diffs are small betw individuals of same species
small diff in DNA base sequences between individual organisms within a species population = genetic variation
genetic variation is…
transferred from one generation to the next and it generates phenotypic variation within a species population
what processes cause genetic variation (result in new combo of alleles in gamete/individual)
independent assortment of homologous chromosomes during M1
crossing over of non-sister chromatids during P1
random fusion of gametes during fertilisation
mutation results in…
generation of new alleles
new allele may be advantageous, disadvantageous or have no apparent effect on phenotype (because genetic code is degenerate)
new alleles are not always seen in the individual that they first occur in
they can remain hidden (not expressed) within a population for several generations before they contribute to phenotypic variation
genes have varying effect on organisms phenotype
may be affected by a single gene or by several
impact that gene has on phenotype may be large, small, and/or additive
mechanism and consequence of independent assortment of homologous chromosomes during M1
random alignment of chromosomes results in different combinations of chromosomes and different allele combinations in each gamete resulting in genetic variation between gametes
mechanism and consequence of crossing over of non-sister chromatids during P1
exchange of genetic material between non-sister chromatids leads to new combinations of alleles on chromosomes. can also break the linkage between genes resulting in genetic variation between gametes
mechanism and consequence of random fusion of gametes during fertilisation
any male gamete can fuse with any female gamete (random mating in a species population) resulting in genetic variation between zygotes and resulting individuals
mechanism and consequence of mumtation
random change in the DNA base sequence results in the generation of a new allele. mutation must exist within gametes for it to be passed on to future generations resulting in genetic variation between individuals within a species population
how can the environment an organism lives in impact phenotype
different conditions affect how organisms grow and develop
length of sunlight hours (seasonal)
supply of nutrients (food)
availability of water
temperature range
oxygen levels
can environmental pressure caused variation be inherited
no, only alterations to genetic component of gametes will ever be inherited
variation
differences that exist between at individuals of a species (intraspecific variation)
why is there variation in phenotypes
due to qualitative or quantitative differences
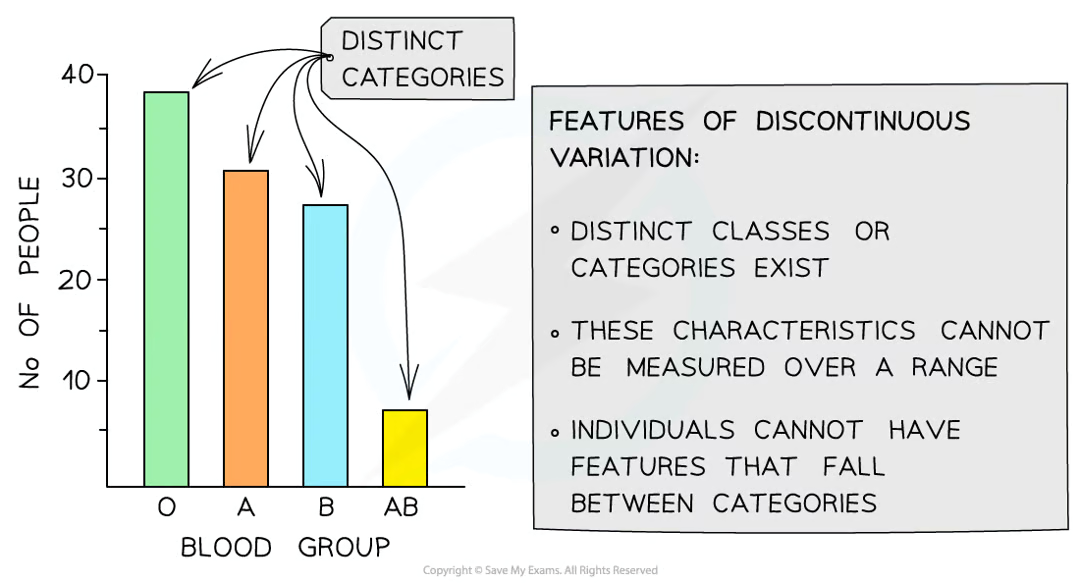
qualitative differences in phenotypes of individuals of population =
discontinuous variation
qualitative differences fall into ______ and _______ categories with __ ____________
discrete and distinguishable categories, usually w no intermediates (feature cant fall in between categories e.g. 4 possible ABO groups and person only has one)
is it hard to identify discontinuous variation
no, it is easy when its present in a table or graph due to the distinct categories that exist when data is plotted for particular characteristics
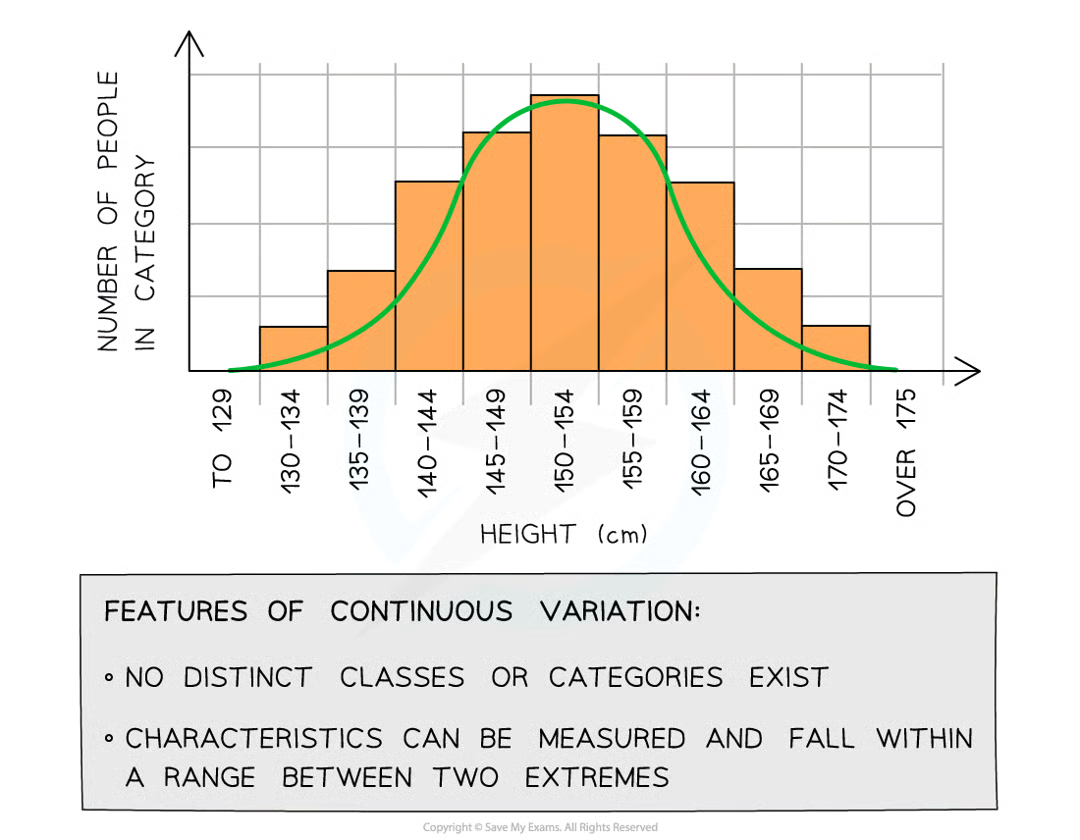
continuous variation
occurs when there are quantitative differences in the phenotypes of individuals within a population for particular characteristics
do quantitative differences fall into discrete categories
no, a range of values exists between 2 extremes within which the phenotype will fall (e.g. mass/height of human). the lack of categories and presence of range of values can be used to identify continuous variation when presented in table/graph
discontinuous variation refers to
the differences between individuals of a species where the differences are qualitative (categoric)
continuous variation refers to
the differences between individuals of a species where the differences are quantitative (measurable)
each type of variation can be explained by…
genetic and/or environmental factors
genetic basis of discontinuous variation
solely due to genetic facgtors
environment has no direct effect
at the genetic level diff genes have diff effects on phenotype and diff alleles at single gene locus have large effect on phenotype (diploid orgs will inherit 2 alleles of each gene and can be same or diff alleles)
e.g. f8 gene coding for blood clotting protein factor viii - diff alleles at f8 gene locus dictate whether or not normal factor viii is produced and whether haemophilia
genetic basis of continuous variation
caused by interaction between genetics and environment
phenotype = genotype + environment
at genetic level diff alleles at single locus have small effect on phenotype and diff genes can have same effect on phenotype and these have an additive effect. if large no. genes have combined effect on phenotype = polygenes
t-test
statistical test used to compare means of 2 sets of data and determine whether significantly diff or not
for t test what must sets of data be
follow roughly normal distribution
continuous
standard deviations should be approx equal
what must be calculated for each data set before the t test can be carried out
standard deviation

null hypothesis
this is a statement of what we would expect if there is no significant difference between 2 means and that any differences seen are due to chance
when is the null hypothesis rejected
if there is a statistically significant difference between the means of two sets of data, then the observation is not down to chance and nh rejected
steps to calculate t-test
calc mean for each data set
calc standard deviation for each set of data
square the standard deviation and divide by n (no. observations) in each sample for both samples
add values from step 3 together and take square root
divide diff between 2 means w value calc in step 4 to get the value
calc degrees of freedom for whole data set (v = (n1-1) + (n2-1)
look at table that relates t values to probability that diff between data sets is due to chance to find where t value for degrees of freedom v calculated lies
the greater the t value calc (for any dof) the lower the probability of chance causing any significant difference between the two sample means.
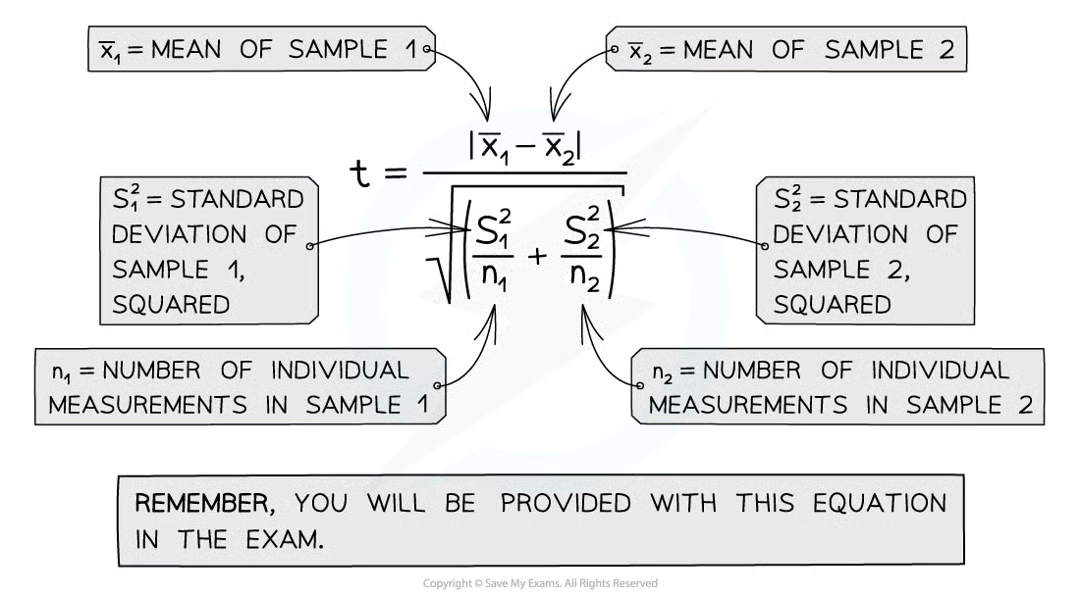
if t value is greater than the critical value (0.05)
any diff between the means of the two data sets is said to be statistically significant = there’s less than 5% probability that any difference is due to chance, null hypothesis rejected
if t value is less than critical value (0.05)
then no sig diff between mean of 2 data sets. probability that any diff is due to chance is higher than 5% so null hypothesis is accepted
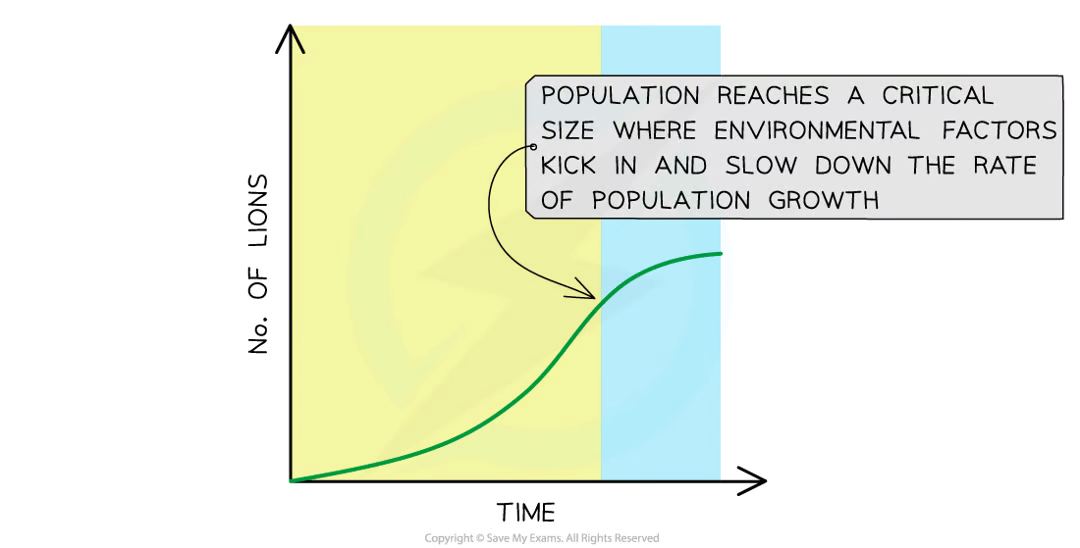
when would a population experience exponential growth
if the offspring for every individual survived to adulthood and reproduced then the pop = exp growth
only happens when no environmental factors or pop checks are acting on pop (e.g. lots of resources and no disease)
one well-known but rare example of expo growth = introduction of european rabbits to aussie
rabbits had an abundance of resources little comp and no predators so increased rapidly = pest
in reality, several env factors prevent every individual in a pop from making it to adulthood and reproducing
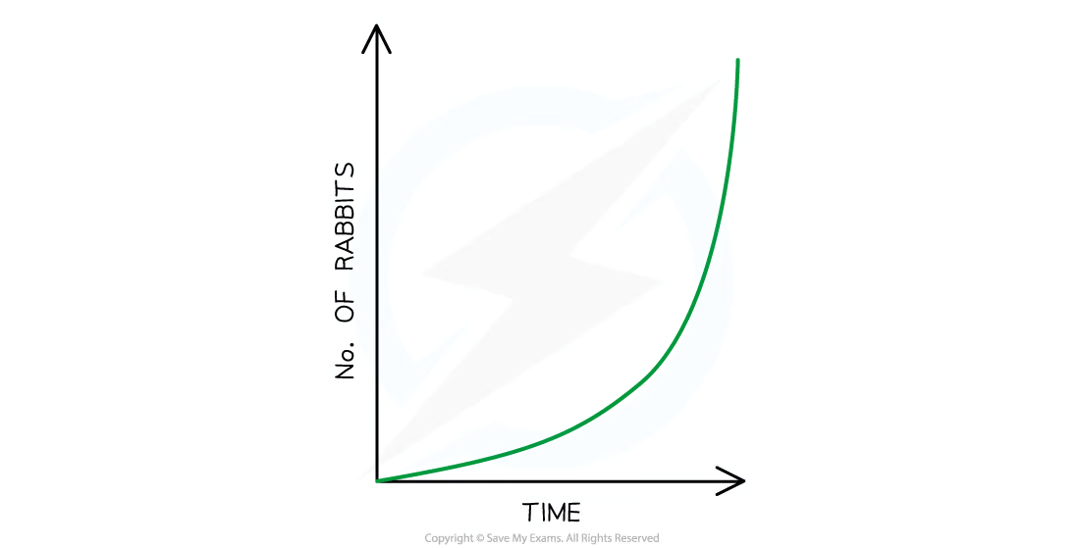
how do env factors limit pop sizes
by reducing rate of pop growth whenever a pop reaches a certain size
env types of factors
biotic
abiotic
biotic
involve other living organisms e.g. predation, competition for resources, disease
abiotic
non living parts of environment e.g. light availabiltiy, soil pH, water supply
no offspring produced is much ____ than…
higher than no. individuals that make it to adulthood
environmental factors that limit pop growth rate (african lions in wild)
comp for food - limited supply of prey. others also hunt. if no food = starve
comp for mate - more females so males compete. they fight = death/injury. loss = no mate so no offspring
supply of water - can be v arid during dry season. far water sources. die dehydration if dries up.
temp - extreme heat = overheat and die. stops from hunting so no food.
= decrease in pop growth
variation
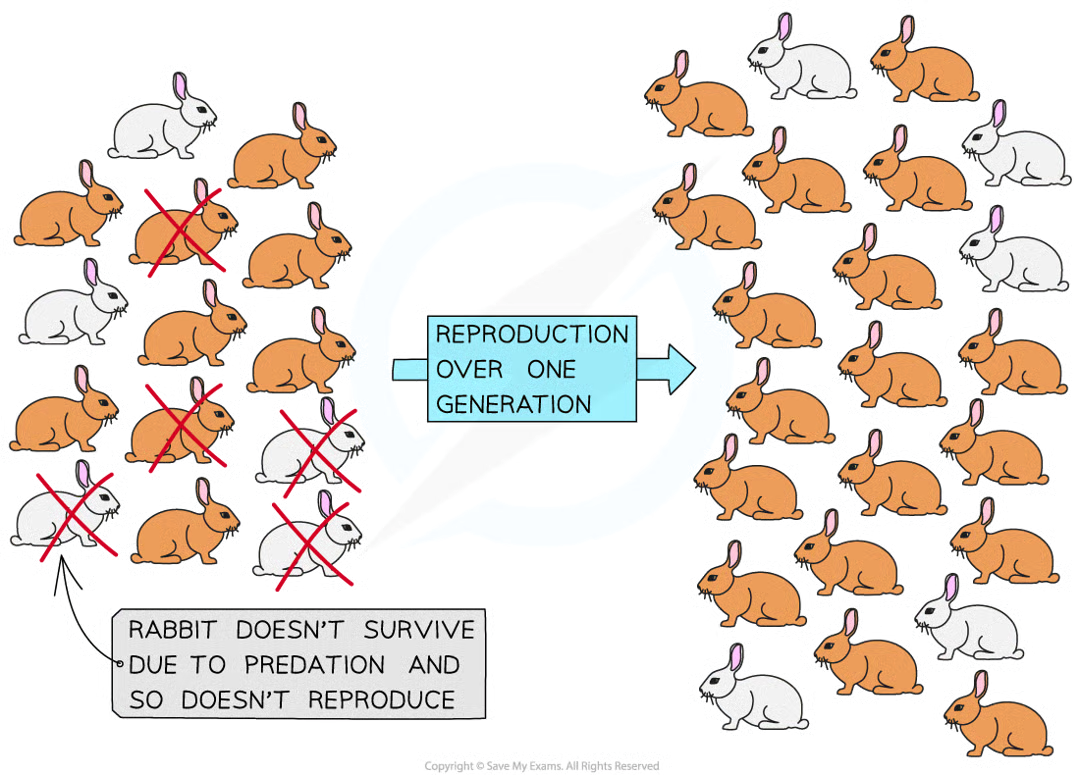
exists within a species pop meaning some individuals within pop posses diff phenotypes bc of genetic variation in alleles they possess. members of same species have same genes
selection pressure
increases chance of individual w specific phenotype surviving and reproducing others and env factors can act as this.
fitness
ability to survive and pass on alleles to offspring. orgs w higher fitness = favoured/possess adaptations that make them better suited to environment
natural selection
process by which individuals w fitter phenotype are more likely to survive and pass on their alleles to their offspring so that the advantageous alleles increase in freq. over time and generations. when selection pressures act over several generation of species they effect frequency of alleles.
3 types of selection
stabilising
directional
disruptive
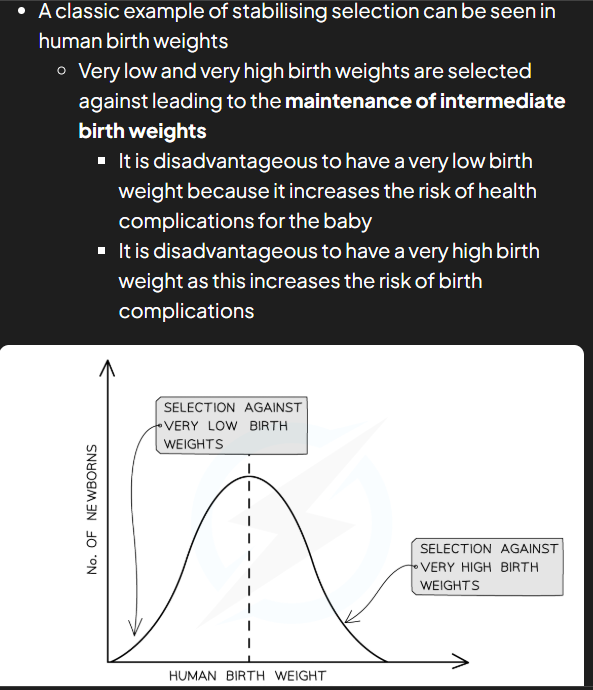
stabilising selection
natural selection that keeps allele frequencies constant over generations meaning that allele frequencies stay constant unless there’s a change in environment e.g. human birth weights
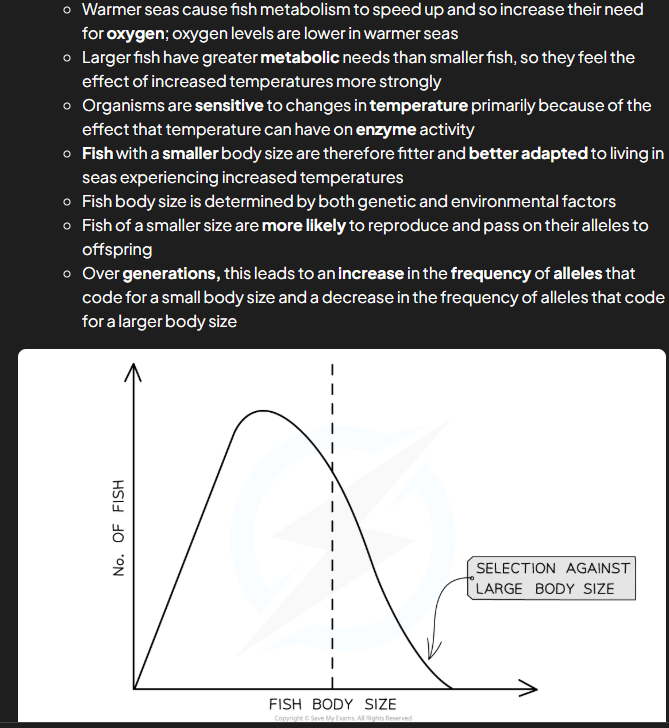
directional selection
nautral selection that produces a gradual change in allele frequencies over several generations usually when there’s a change in environment or new selection pressures leading to certain alleles becoming advantageous e.g. fish size (smaller in hotter temps)
disruptive selection
natural selection that maintains high frequencies of two diff sets of alleles e.g. individuals with intermediate phenotypes/alleles are selected against e.g. galapagos birds
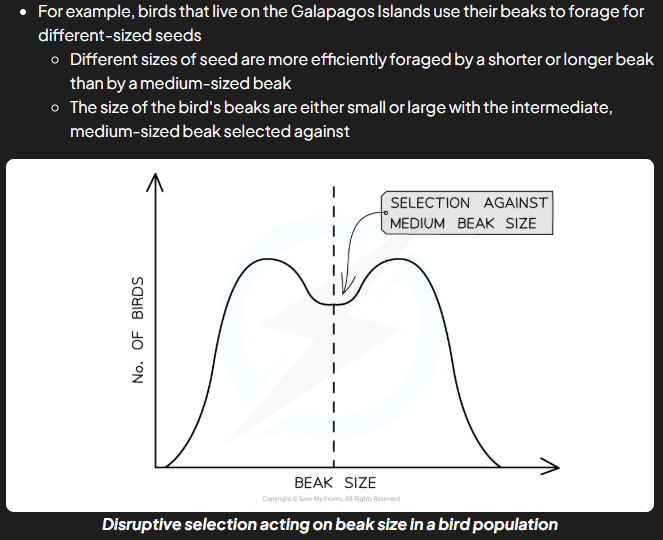
what does disruptive selection maintain
polymorphism: the continued existence of 2 or mroe distinct phenotypes in species (variation)
genetic drift
when allele frequencies change bc of chance
other processes that can cause allele freq changes due to chance:
founder effect
bottleneck effect
natural selection
when new allele arises in pop or change in env occurs directional selec can happen producing gradual change in allele freq over sev generations
always phenotypic variation
selection pressure
some indi may have phenotype that aids survival in presence of selec pressure produced by particular alleles
indi w favored phenotype fitter so reproduce and pass on to offspring
those who dont have less likely to suurvive
overtime and generations, freq of advantageous increases and other decreases
genetic drift
when pop v small, chance can affect which alleles get passed on to next gen
meiosis = haploid gametes so fert only passes on half alleles of individual
half that gets passed is result of random fertilisation and other half may not make it to next gen
overtime some alleles lost or passed purely by chance
effects smaller pops e.g. coin toss 10 times = heads wont come but if 100 less likely etc
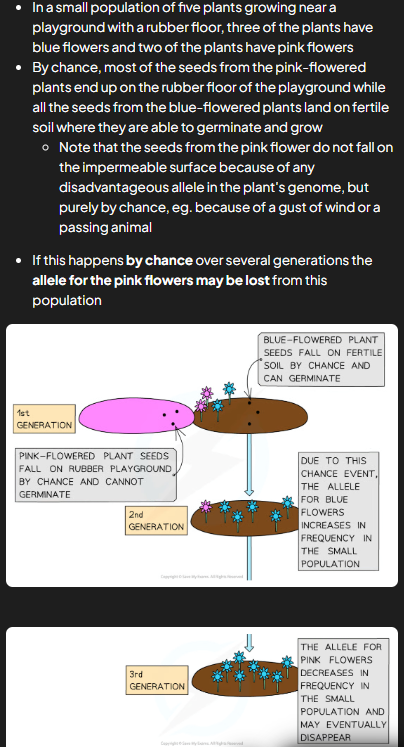
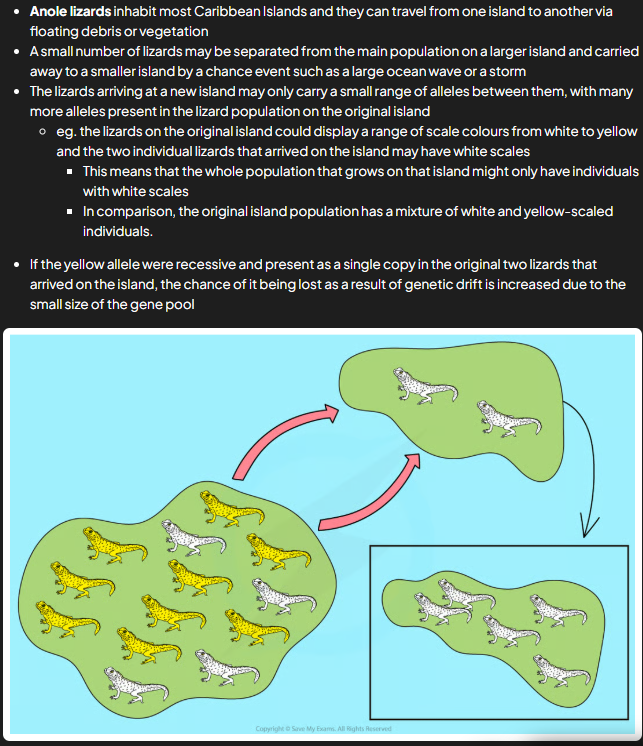
founder effect
occurs when small no. individuals from large parent pop start new pop and can come about as result of chance e.g. storm separates small grp of indi from main pop
as new pop made up of only few indi from og pop, only some of total alleles from parent pop present e.g. not all gene pool present in smaller pop
bc pop results from founder effect is very small, more susceptible to effects of gen drift
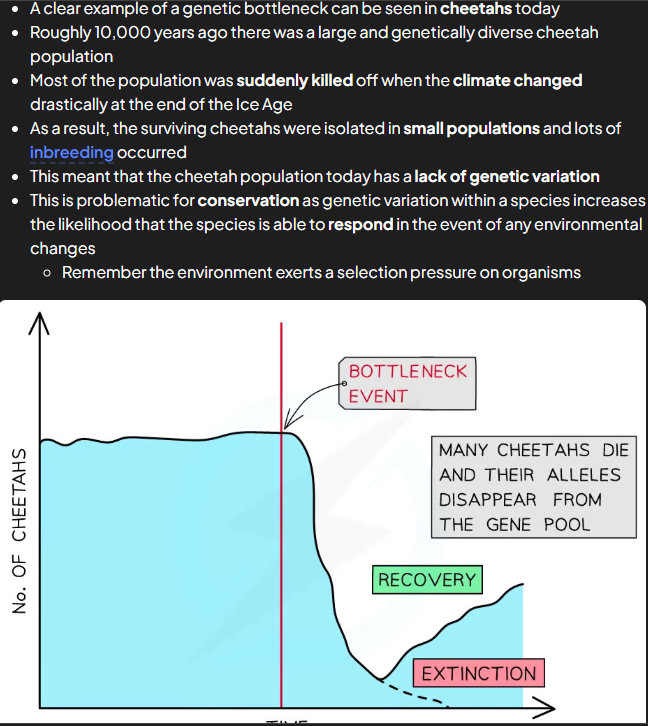
bottleneck effect
similar to founder and occurs when prev large pops suffers dramatic fall in numbers
major env event can greatly reduce no. indi in pop = reduced gen diversity in pop as alleles lost
surviving indis end up breeding and reproducing w close relatives
4 processes affecting allele freq and result
natural selec - selec pressures produce gradual change in allele freq over sev gen
founder effect - changes in allele freq occur in diff direction for newly isolated small pop in comparison to larger parent pop due to chance
gen drift - gradual change in allele freq in small pop due to chance not natural selec
bottleneck effect - reduction in gene pool of pop due to dramatic decrease in pop size
antibiotics
chemical substances that inhibit or kill bacterial cells w little or no harm to human tissue. derived from naturally occuring substances that are harmful to prok cells but dont affect euk cells (aids bodys immune system in fighting bac infection)
bactericidal vs bacteriostatic
cidal = kill
static = inhibit growth process
how do antibiotics work
target prok features but can affect both pathogenic and mutualistic bac living on body
bacterial resistance
all bac have genetic diversity and indi bac may have alleles that confer resistance to effects of antibiotic. alleles gen thru random mutation, not by anitbiotic use but use exerts selec pressure that can = increase in freq
bacteria dna
single loop of dna w only one copy of each gene so when new allele arises its immediately displayed in phenotype
when antiobitic present:
individuals w allele for antibiotic res have massive selec adv so more likley to survive, reprod and pass genome including resistance alleles on
those without alleles less likely to survive and reprod
over sev gens, entire pop of bac may be resistant
staphylococcus
have resistant strains
due to rapid reprod rate of bacterial (gens of 20-30 mins for some species), single res bacterium can prod 10,000 mil resistant descendants in a day
can strains of bac be resistant to multiple antibiotics
yes, v difficult to treat and hard to eradicate.
hardy weinberg principle
used to predit allele freq in pop only under certain conditions in pops where:
no natural selection happening
no migration in or out of pop
mating random
pop large
no mutations
if phenotype of trait in pop is determined by single gene w only 2 alleles then pop will consist of indi w 3 possible genotypes:
homozygous dom (BB)
heterozyg (Bb)
homozygous rec (bb)
hardy weinberg equation freq of alleles
frequencies represented as proportions of pop (no. out of 1)
freq of alleles can be rep; this is prop of all of alleles in pop that are of particular form
p = dominant
q = recessive
only 2 alleles at single gene locus for a phenotypic trait in pop
p+q = 1
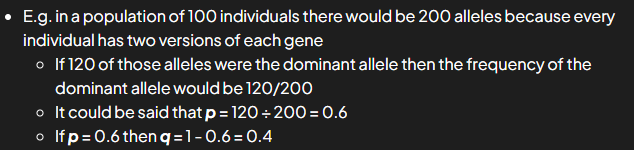
hardy weinberg equation freq of genotypes
proportion of all individuals w particular genome
freq of homozygous dominant individuals p²
freq of heterozygous individuals 2pq
freq of homozygous recessive = q²
equation: p² + q² + 2pq = 1

artificial selection
process by which humans choose organisms w desirable traits and selective breed them together to enhance the expression of these desirable traits over time and many generations also known as selective breeding e.g. increased milk yield from cattle, faster racehorses, disease-resistant crops
limits to how extreme trait can become
why do breeders accidentally enhance other traits genetically linked to desirable
individuals are selected based on phenotypes. can negatively affect orgs health
6 principles of selective breeding
pop shows phenotypic variation - individuals w diff phenotypes/traits
breeder selects indi w desired phenotype
another individual w desired phenotype is selected. two selected individuals shldnt be closely related
two selec indi bred tg
offspring produced reach maturity and are then tested for desirable trait. those display desired phenotype to greatest degree selected for further breeding
process continues for many gens: best indi from offspring chosen for breeding until all offspring display desirable trait
selective breeding in horseracing industry. 3 phenotypes: +
good at sprinting short dist
good endurance over long dist
all-rouder
how do horsebreeders breed horse for sprinting
select fastest sprinting female and male horse
breed two and allow offspring to grow and test sprinting speeds to find fastest
breeder could use for racing or continue breeding w other fast sprinters
= all fast offspring
why is most selective breeding done
to increase yield of saleable prod, not done w orgs survival or health in mind so can = unfit orgs and other traits can be accidentally enhanced e.g. disease resistance in wheat and rice varieties, hybridisation in maize, milk yield cattle
disease resistance in wheat and rice
wheat can be badly affected by fungal diseases e.g. fusarium that causes head blight in wheat plants
fungal diseases highly problematic for farmers as destroy wheat plant and reduce crop yield
by using selective breeding to intorduce a fungus-resistant allele from another species of wheat, hybrid wheat plants are not susceptible to infection, and so yield increases
introducing allele into crop pop can take many gens and collab w researchers and plant breeders
rice is another, e.g. bacterial blight and rice blast from magnaporthe fungus reducing yield
scientists working to create varieties of rice plants resistant to several bac and fungal diseases
inbreeding and hybridisation in maize
maize/corn was heavily inbred in past resulting in small and weaker plants w less vigour
inbreeding depression which:
increases chance of harmfulr ecessive alleles combining in indi and being expressed in phenotype
increases homozygosity in indi paired alleles at loci are identical
leads to decreased growth and survivability
farmer can prevent by outbreeding (breeding indi not closely related). will result in taller and heatlhier maize plants. decreases chance of harmful recessive alleles combining in indi being expressed in phenotype. increased heterozygosity and = hybrid vigour and increased growht and survivability therefore yield
unifromity is important when growing crop as if outbreeding carried too randomly = variation so farmer needs plants to ripen and be similar hypes. this is done by buying sets of homozygous seeds from specialised companies and cross to produce f1 gen. constant for desirable traits.
improving milk yield in cattle
female cows that have highest milk yield get crossed w male bulls related to high yield females = greater milks yields and econ bens for farmers
doesnt take into account orgs survival as focuses on extreme characteristics
little though given to other traits, selectively bred indi more prone to ailments e.g. mastitis (inflam of udder), milk fever, and lameness
species
group of organisms that are able to interbreed and produce fertile offspring. members of one species are repdocutively isolated rfom members of another species.
individuals of the same species have
similar behavioural, morphological (structural) and physiological (metabolic) features
gene pool
collection of genes within an interbreeding population and sum of all alleles at all of the loci within the genes of pop of a single species or a population
3 reasons gene pool changes over time:
natural selection
genetic drift
founder effect
evolution
takes long but can be quicker in bacteria if short gen time
when gene pool within species pop changes sufficiently over time, the characteristics of species will also change. change so great that new species forms.
for a population to evolve into separate species it must be…
genetically and reproductively isolated from pre existing species conditions
reproductive isolation
can occur due to mutations that lead to incompatibility of gametes or sex organs, or diff in breeding behavior
when two pops are reproductively isolated, they are also…
genetically isolated so they dont exchange genes w each other in prod of offspring
speciation
changes in allele freq of isolated pop not shared so they evolvev independently of each other = 2 groups that cannot interbreed and are separate species
how can evolutionary relationships between species be shown
dna found in nucleus, mitoch and chloroplasts of cells can be sequenced
differences between nucleotide sequences/dna of diff provide what info: 2
more similar sequence = more closely related species
two groups of orgs w/ v similar dna will have separated into separate species more recently than 2 groups w less similarity in their dna sequences
dna sequence analysis and comparison can do what
used to create family trees/phylogenetic trees that show evolutionary relationships between species

how is dna analysis done
dna extracted from nuclei of cells taken from org (can be from blood or skin or fossils)
extracted dna processed, analysed, base sequence obtained
base sequence is compared to that of other orgs determine evolutionary relationships (more similarities in dna = more closely related diff species)
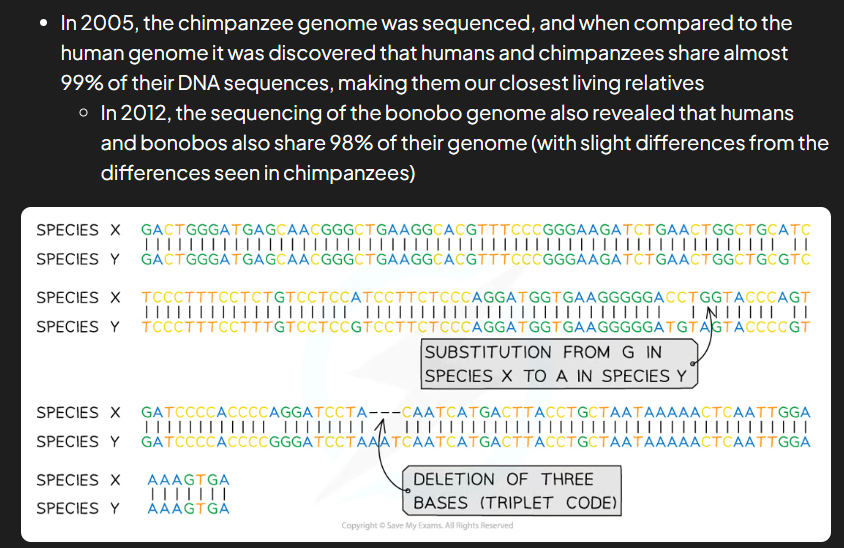
when analysing dna must remember: 2
zygote only contains mitoch of egg and none from sperm so only maternal mitochondrial dna is present in zygote
no crossing over that occurs in mtDNA so base sequence can only change by mutation
what has allowed scientists to research origins of species, genetic drift, and migration eventsq
lack of crossing over in mtDNA.
this is how estimated when humans first lived (200,000 years ago in Africa)
estimation of date relies on molecular clock theory which assumes constant rate of mutation over time
greater no. diff between nucleotide sequences, longer ago common ancestor existed
molecular clock calibrated w fossils and carbon dating
fossil of known species is carbon-dated to estimate how long ago org lived
mtDNA of species then used as baseline for comparison w mtDNA of other species
can only maternal mitoch dna be passed on or inherited by zygote
(yes for exma but curently research showing paternal mDNA)
evolution causes…
speciation: the formation of new species from pre-existing species over time as a result of changes to gene pools from gen to gen
2 different situations when speciation can take place:
2 grps within species separated by geographic barrier
2 grps of species are reproductively isolated but still living in same area (experiencing similar env selec pressure)
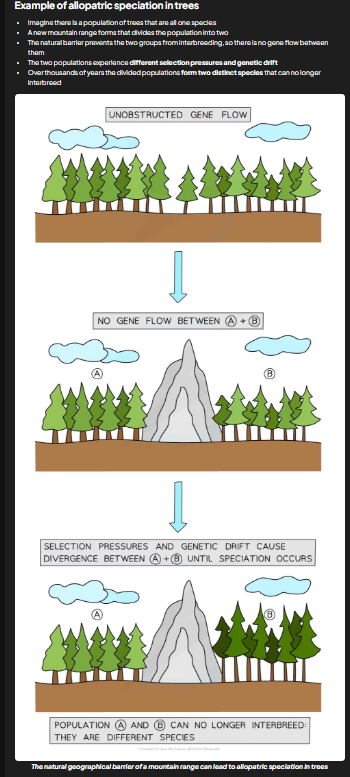
allopatric speciation
result of geographical isolation
most common
species pop splits into one or more groups which become separated from each other by geographical barriers e.g. mountain, body of water, motorway
separation = 2 pops of same species isolated so no genetic exchange occuring
if sufficient selection pressure/genetic drift acting to change the gene pools within both populations then eventually these population will diverge and form separate species
changes in alleles/genes of each pop affect phenotypes present in both pops
2 pops may begin to differ physiologically, behaviorally, and morphologically
example in pic
sympatric speciation
takes place w no geographical barrier
group of same species living in same place but in order for speciation to take place there must exist 2 pops within that group and no gene flow occurs between
something has to split/separate population
ecological separation: populations are separated bc they live in diff environments within same area e.g. soil pH can differ greatly in diff areas so effects flowering and growth
behavioural separation: pops separated bc diff behaviors e.g. feeding, comms, social behavior
example in pic
