BIOL121: Lecture 6-7
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
88 Terms
What is the central dogma?
genes (DNA) > transcript (RNA) > protein (function)
What step of gene expression is transcription?
first
T/F: Transcription is guided by transcribed DNA regulatory sequences upstream of the gene
false
guided by un-transcribed DNA regulatory sequences
What is the core enzyme?
RNA polymerase for RNA synthesis
What are the subunits of RNA polymerase?
alpha alpha beta beta
T/F: Sigma factor is needed for initiation and elongation of RNA synthesis
false
only initiation not elongation
How does the sigma factor recognize promoters?
recognizing promoters by binding to 10 and 35 regions of genes
Why is the sigma factor important?
guides core enzyme/RNA polymerase to promoter to initiate transcription
What is the core enzyme plus sigma factor called?
holoenzyme
Bacteria utilize multiple sigma factors to regulate transcription of () genes in response to () environmental conditions
different x2
() can be seen as the “housekeeping” sigma factor
sigma70
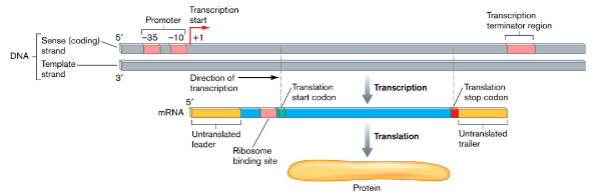
Initiation of Transcription
() binds to DNA
() recruits () and scans for ()
() unwinds DNA at ()
() is released
sigma factor
sigma factor, core enzyme, promotor region
core enzyme, promotor (open complex)
sigma factor
T/F: Core polymerase synthesizes RNA strand 5’ to 3’
true
T/F: Added base is complementary to template strand
true
mRNA has same sequence as () strand but uses U instead of T
non-template
Rho () termination forms a stem loop causing the mRNA to break off the DNA and release the polymerase
independent
Rho () termination has the helicase bind to the mRNA, pulls itself to the paused RNA polymerase and breaks the polymerase
dependent
In Rho independent termination the () sequence forms the stem loop and the () beneath the pause site is unstable, causing the mRNA to break off the DNA
GC
UA
What are the 4 types of RNA?
mRNA - messenger
rRNA - ribosomal
tRNA - transfer; carry information from DNA to protein
sRNA - small RNAs; regulate stability or translation of specific mRNAs into protein
() is the decoding of RNA into protein
translation
() is the reading of a DNA template to make an RNA copy
transcription
Open reading frame is contained within mRNA and located between () codon and () codon
translation start
stop
T/F: Each transcript has one reading frame
false
three possible
Stop codon in () frame as start codon
same
tRNAs have () structure and () which are base pair to codons in mRNA
cloverleaf
anticodon
() attach amino acid aa to tRNA to charge the tRNA
aminoacyl-tRNA transferases
T/F: Ribosomes are composed of 2 subunits: 1 small and 1 large
True
Ribosomes bind to () mRNA and () tRNAs
1
3
Transcription and translation are () in prokaryotes
coupled
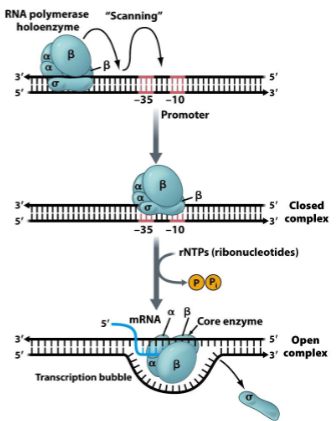
Translation Initiation
Ribosome binding site () on mRNA allows binding to () subunit
() brings “bottom” of ribosome and mRNA together
() blocks () site
() escorts () to start codon
“top” () of ribosome docks to “bottom” () subunit
shine-dalgarno sequence, 30s
IF3
IF1, A
IF2, formylmethionine tRNA
50s, 30s
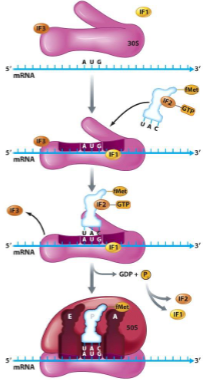
Translation Elongation
() binds to () site
() formed between new aa and () in the P site
Ribosome shifts down mRNA () codon
aminoacyl-tRNA, A
peptide bond, growing peptide chain
one
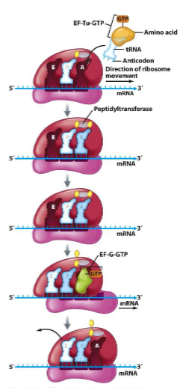
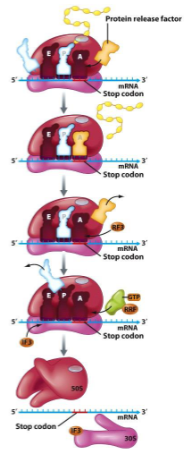
Translation Termination
() on mRNA enters () site
() factor enters A site
() activated and releases () protein
() factor and () enter A site and ribosome falls apart
stop, A
protein releasing
peptidyltransferase, complete
ribosome recycling, EF-G
Microbial genomes are () as a result of heavy horizontal gene transfer, (), and a variety of () and () strategies.
mosaic
recombination
mutagenic
DNA repair
Microbial genomes have undergone extensive gene ()
loss and gain
T/F: DNA sequences are static
false
not static
T/F: Mutations of single bases happens quickly
false
slow, accumulated through time
Large insertions of sequences are called () and transferred from other species is called ()
genetic islands
horizontal gene transfer
4 reasons why DNA sequences are not static
mutations of single bases
large deletions
large insertion of sequence
plasmids
Plasmids are () DNA, usually circular and transferred by () and ()
extrachromosomal
transformation
conjugation
What are the size and copy number of plasmids?
several thousand to million bases
0-100 per cell
Why is it important plasmids have their own origin of replication?
important for bacterial adaptation to changing environments
ex antibiotic resistance
5 plasmid advantages under special conditions:
antibiotic resistance genes
genes encoding resistance to toxic metals
genes encoding proteins to metabolize rare food sources
virulence genes to allow pathogenesis
genes to allow symbiosis
What are the 3 transfer processes of horizontal gene transfer?
transformation
free DNA taken up from environment and integrated into genome
conjugation
cell-cell contact
transduction
DNA transfer by bacteriophage
Who first demonstrated discovery of transformation?
Griffith, Avery, and MacLeod
R strain and heat killed S strain
Cell needs to be () in order for gene transformation to happen
competent
In gene transformation, () in Gram positive bacteria takes up DNA, bacteria secretes (), and accumulation of it induces assembly of () located in cell membrane
translocasome
competent factor
translocasome
In gene transformation, () induces (), which is the ability of cells to take up foreign DNA
starvation/stress
competence
() is the transfer of DNA from one bacterium to another, following cell-to-cell contact, often called () and initiated by a () protruding from donor cell
conjugation
bacterial sex
special pilus
Sex pilus is type () secretion system
4
Gene Transfer: Conjugation
() proteins encoded on fertility plasmid or ()
Fertility plasmid or () is (), as it makes its own transfer machinery
In recipient cell, transferred DNA receives () and becomes ()
() cells become (), able to transfer DNA
Recipient is always ()
pilus, F factor
F factor, self-transmissible
a copy of F factor, F+
Female, male
F-
Some bacteria can transfer genes across ().
Agrobacterium tumefaciens which causes crown gall disease contains tumor-inducing plasmid that can be transferred via () to plant cells.
biological domains
conjugation
() is the process in which bacteriophages carry host DNA from one cell to another.
This occurs () as offshoot of phage life cycle.
transduction
accidentally
() transduction can transfer any gene from donor to recipient and () can transfer only a few closely linked genes between cells
generalized
specialized
Gene Transfer: Transduction
Viruses inject () into cell
At end of life cycle, viral DNA packaged into () before ()
Sometimes () by mistake
Virus carries ()
Transfers DNA to new host and brings DNA from previous host to new host
viral DNA
viral capsid, cell lysis
package bacterial DNA
host DNA - transducing particles
In specialized transduction, () is when some viruses can integrate their genome into bacterial chromosome at att sites.
When entering (), bacterial genes () to viral () sites are sometimes mistakenly picked.
lysogeny
lytic cycle
adjacent
attachment
Most foreign DNA will be degraded by () in recipient cell.
Plasmids will coexist in cell as ().
DNA can incorporate into chromosomal DNA by ().
restriction endonucleases
extrachromosomal DNA
recombination
In order for foreign DNA to incorporate itself into chromosomal DNA by recombination there are 3 requirements:
homologous DNA sequences present
replaces variable-sized section of endogenous DNA
could be used to repair damaged DNA
specific recombination proteins
As a defense against transferred DNA, bacteria adds () groups to its own DNA using () enzymes to protect () sites.
Unless it comes from similar species and has () groups protecting DNA, foreign DNA without DNA () is destroyed.
methyl
matching methylation enzymes
restriction
methyl
methylation
As a defense against transferred DNA, CRISPR is a bacterial () viruses and viral DNA.
When infected, bacteria cut up viral DNA and insert some pieces into () to () for future infections.
immune system against
their own genome
remember
As a defense against transferred DNA, (), which are the little virus bits added to bacterial genome, are transcribed then () monitors the cell for matching invading DNA.
Bacteria () matching DNAs to prevent ().
spacers
Cas9
degrades
viral infection
Mutants generate new () in population and () are heritable changes in nucleic acid bases in genome of organism.
variants
mutations
T/F: Mutations are common and changes the genotype of organism
false
rare
T/F: Mutations become fixed in populations when they are selected for by natural selection
true
What are the 2 types of mutations and what are they caused by?
spontaneous mutations
errors in DNA replication
induced mutations
treating organisms with added mutagens
3 agents causing mutations:
chemical
carcinogens
physical
radiation/heat
biological
insertion of transposons
6 types of mutations:
point mutation
change in single base
transition
purine to purine (A or G)
pyrimidine to pyrimidine (C or T)
transversion
purine to pyrimidine
pyrimidine to purine
insertion/deletion
inversion
DNA flipped
reversion
DNA mutates back to original sequence
() mutation doesn’t change the amino acid because of () of genetic code.
() mutation produces a stop codon.
() mutation leads to amino acid change.
() mutation disrupts the reading frame meaning they aren’t multiples of 3.
silent, degeneracy
nonsense
missense
frameshift
Physical mutagens such as () and () cause formation of toxic () radicals.
Radicals cause two adjacent () bases to () which prevents DNA () and gene ()
UV, ionizing radiation, oxygen
pyrimidine, dimerize, replication, transcription
Chemical mutagens: () are similar structures to natural bases and are incorporated during DNA replication which leads to () because of ().
() changes a bases structure and pairing characteristics.
() distort DNA to induce single nucleotide () or ().
base analogs, point mutation, incorrect base pairing
DNA modifying agents
intercalating agents, insertion, deletion
T/F: Chromosomes sequences are fixed
false
Transposons are () discovered by Barbara McClintock in ().
They are segments of DNA that can () from one place in DNA to another.
They are widely used as () in molecular genetics.
jumping genes, corn
hop
mutagen
T/F: Transposons are limited to bacteria
false
all living things
Transposition: () cuts DNA at () repeat sequences at both ends of transposable element and target sequence of target DNA; and then () the transposable element with target DNA
transposase
inverted
ligates
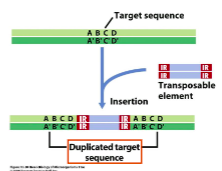
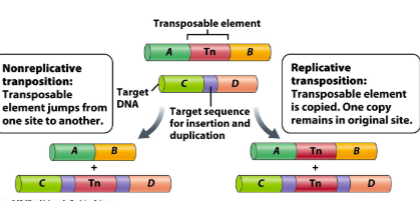
In () transposition, transposable element jumps from one site to another.
In () transposition, transposable element is copied and one copy remains in original site.
nonreplicative
replicative
() examines ability of chemical substance to cause () or strength of () tested.
It is commonly used to determine if chemical has potential to cause cancer
Ames test, mutation, mutagen
Ames test uses () that can not synthesize amino acid ().
Plate bacteria on () with the chemical you are testing for mutagenicity.
bacterial mutant, histidine
defined medium without histidine
On the Ames test plate, more colonies means () mutagens and no growth means () mutagens.
stronger
no
Mutations in DNA repair pathways can lead to () which is the rapid accumulation of mutations.
hypermutation phenotypes
() is the correction of mismatch by DNA polymerase III during DNA replication.
3’ to 5’ () activity that removes () during ()
proofreading
exonuclease, mismatch, polymerization
() is the photolyase enzyme that binds to a pyrimidine dimer caused by UV radiation and cleaves cyclobutane ring
photoreactivation
() repair is the most common damage/mismatch repair and it cuts out () DNA and recopy remaining () strand.
excision
duplex
complementary
Cut, copy, paste mechanism of excision repair
() removes damaged () or ()
() copies template to replace () strand
() seals nick to give ()
enzyme, base, string of nucleotides
DNA polymerase, excised
DNA ligase, intact, repaired DNA
Nucleotide excision repair
recognizes damages that cause () in DNA structure
an () removes patch of () containing damaged bases including ()
distortion
endonuclease, single-stranded DNA, dimers
Base excision repair
recognizes and repairs bases that () DNA structure
recognition at () bases
don’t distort
damaged
Methyl mismatch repair
recognizes () missed by ()
use () as an () for newer strand containing error
mismatches, proofreading
DNA methylation, indicator
() induces SOS regulatory system
() represses expression of SOS genes
() activated by DNA damage to inactivate ()
DNA () are error prone DNA () that repair DNA rapidly, but generate numerous mutations
extensive DNA damage
LexA
RecA, LexA
DNA mutases
() are SUPER bad for bacterial DNA because it causes chromosomal instability, problems with replication, etc
double-strand breaks
Double strand break repair with () ends joining, it doesn’t require (), and () bind broken ends and include () and other proteins recruited to fix break
non-homologous
homology
Ku-family proteins
ligases