Mol Bio Exam I
1/116
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
117 Terms
What are the three main steps of transcription?
Initiation, Elongation, and Termination
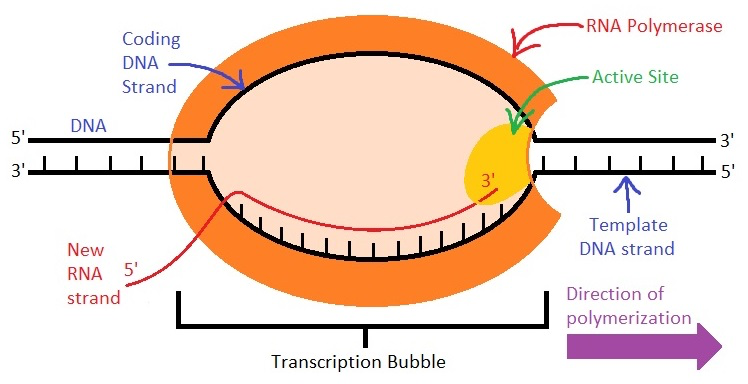
Which direction does RNA polymerase II read the template strand?
from 3' to 5' direction
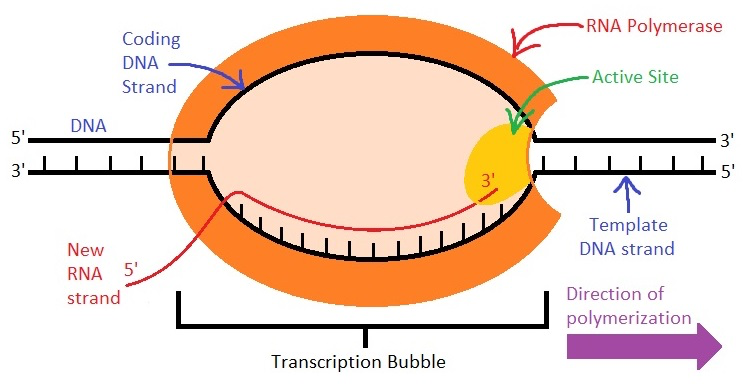
Which direction does RNA pol II synthesize RNA?
in the 5' to 3' direction, adding nucleotides to the 3' end
What is the template strand also called?
non-coding strand, (-) strand, or anti-sense strand
What is the coding strand also called?
the (+) strand or sense strand
How does the RNA sequence compare to the coding strand?
RNA has an identical sequence to the coding strand, except Uracil replaces Thymine
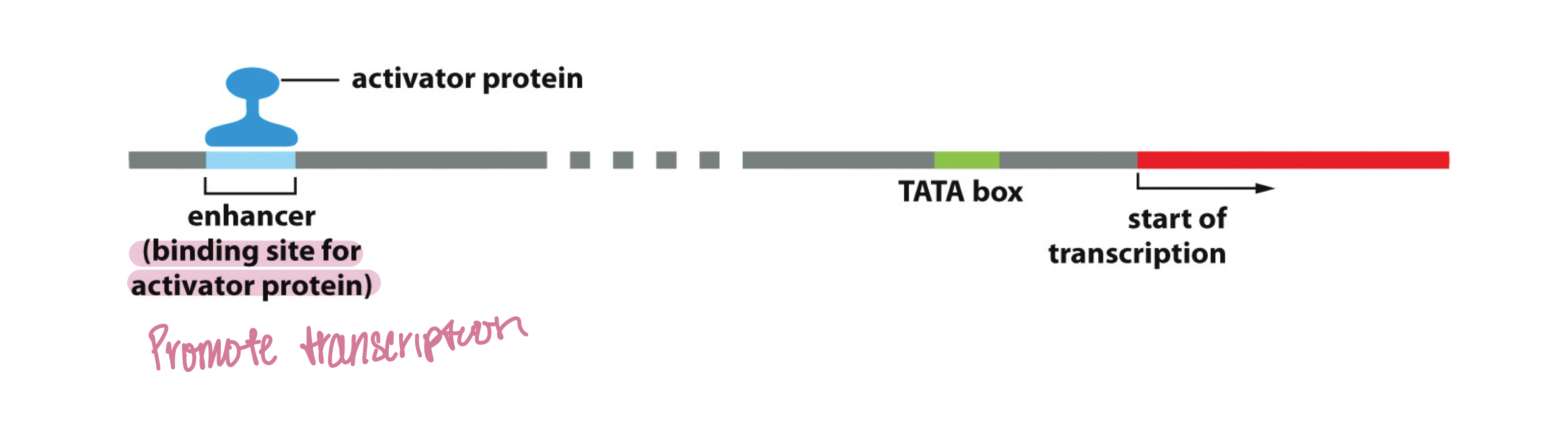
What is the first step in transcription initiation?
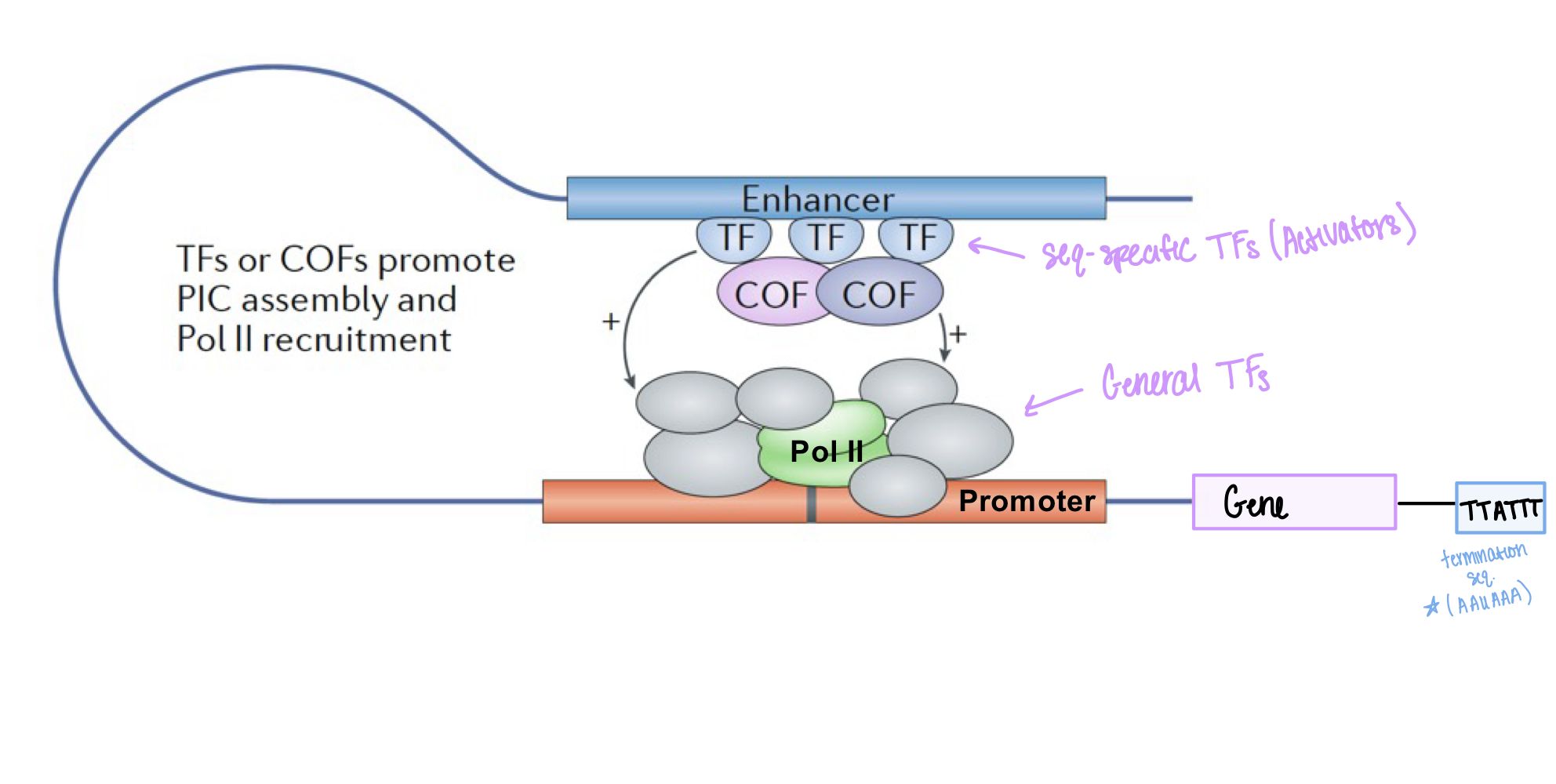
Sequence-specific transcription factors (activators) bind to enhancers using the mediator protein complex
What is the role of the mediator complex in initiation?
facilitates communication between activators and general TFs/pol II, and stimulates phosphorylation of Ser5 in the CTD of pol II
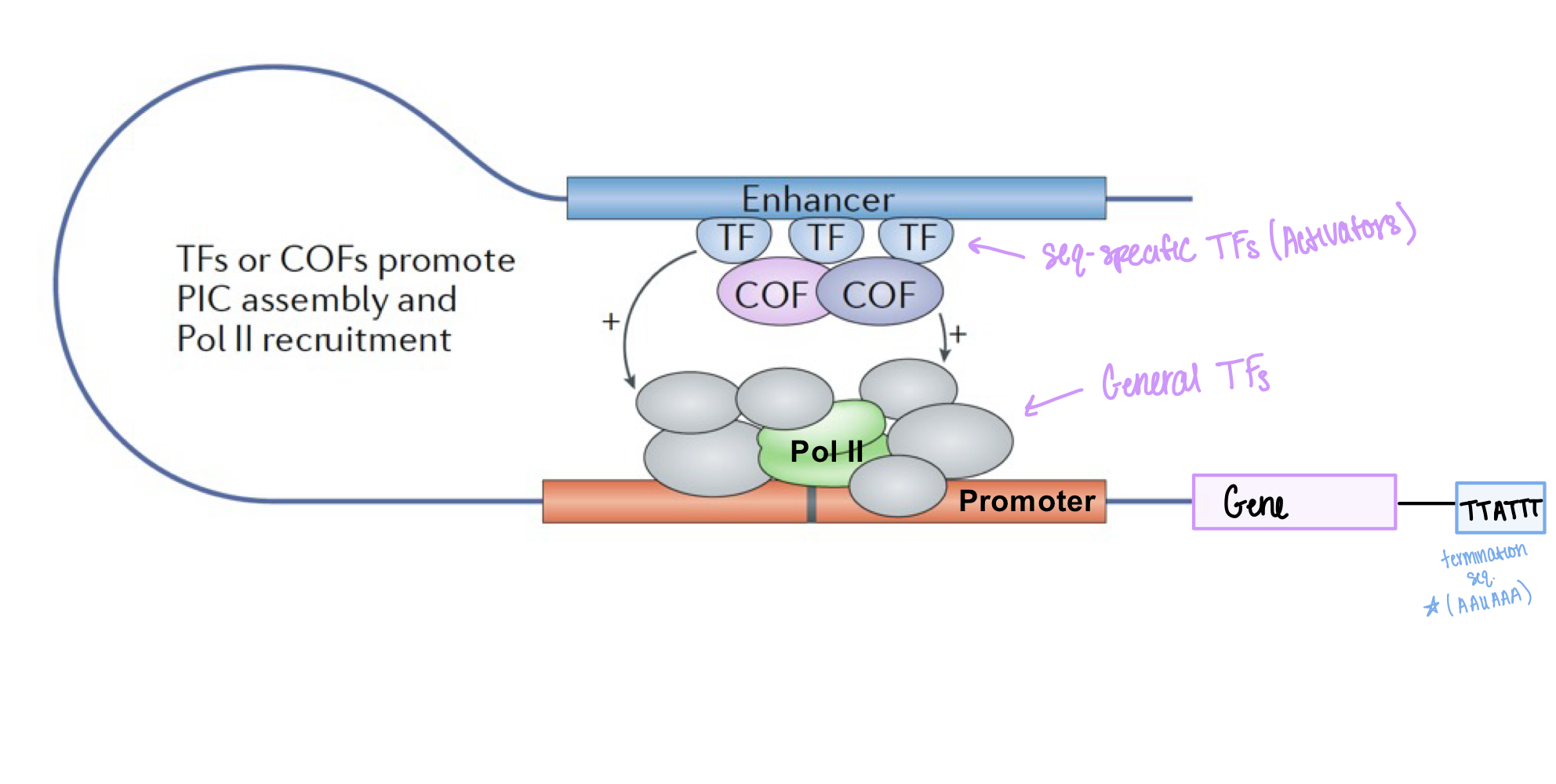
What do cofactors do during initiation?
bind to sequence-specific factors (activators), recruit general TFs, and mediate interactions between sequence-specific TFs and general TFs. They do NOT bind to DNA directly.
Which general transcription factor binds to the TATA box?
TFIID binds to the TATA box region on the promoter
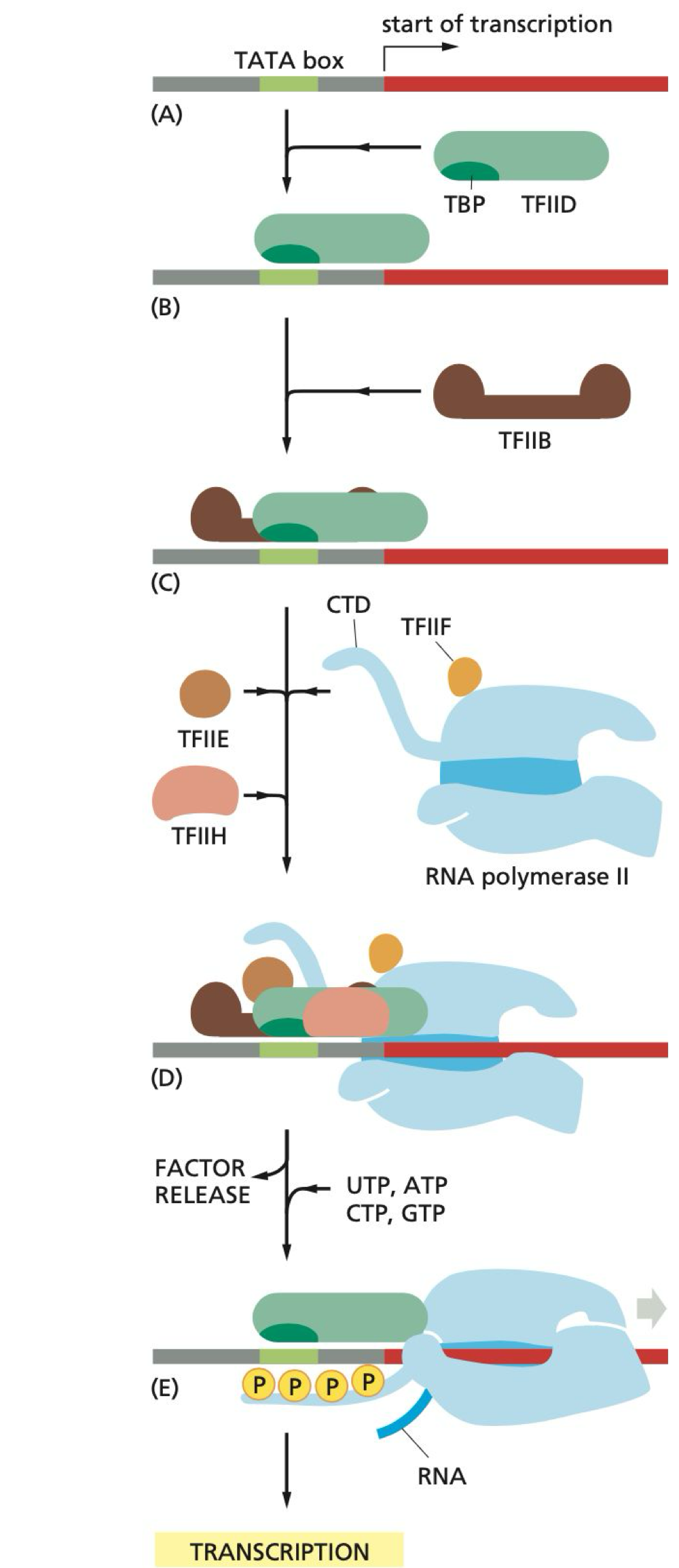
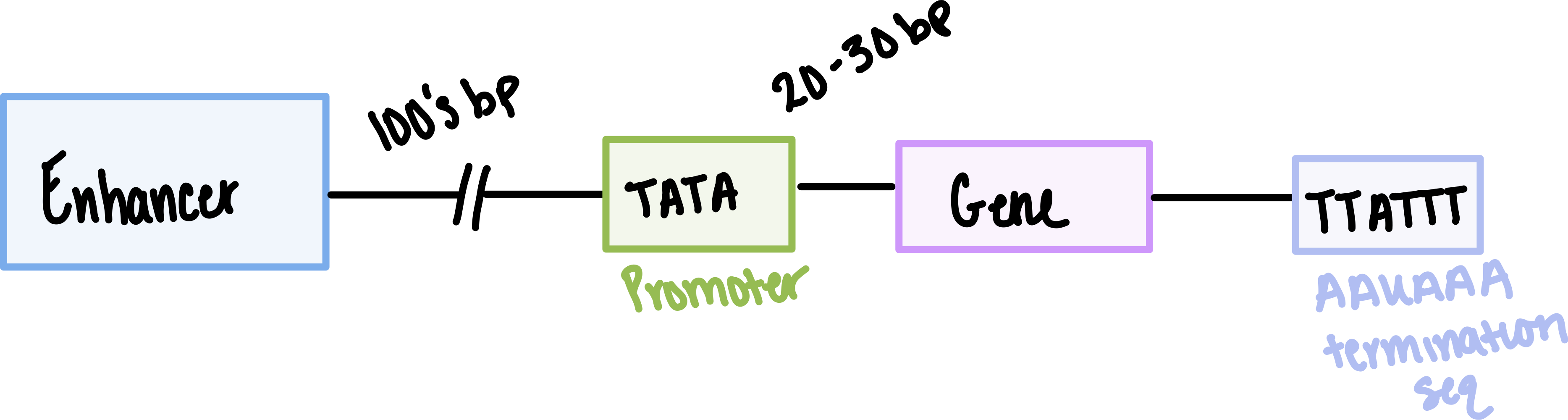
How far is the TATA box from the transcription start site?
The TATA box is 25 bp away from the site of transcription initiation
What does TFIIB do during initiation?
TFIIB recruits RNA pol II to the promoter

What are the functions of TFIIH during initiation?
TFIIH unwinds DNA and phosphorylates Ser5 on the C-terminal domain (CTD) of pol II
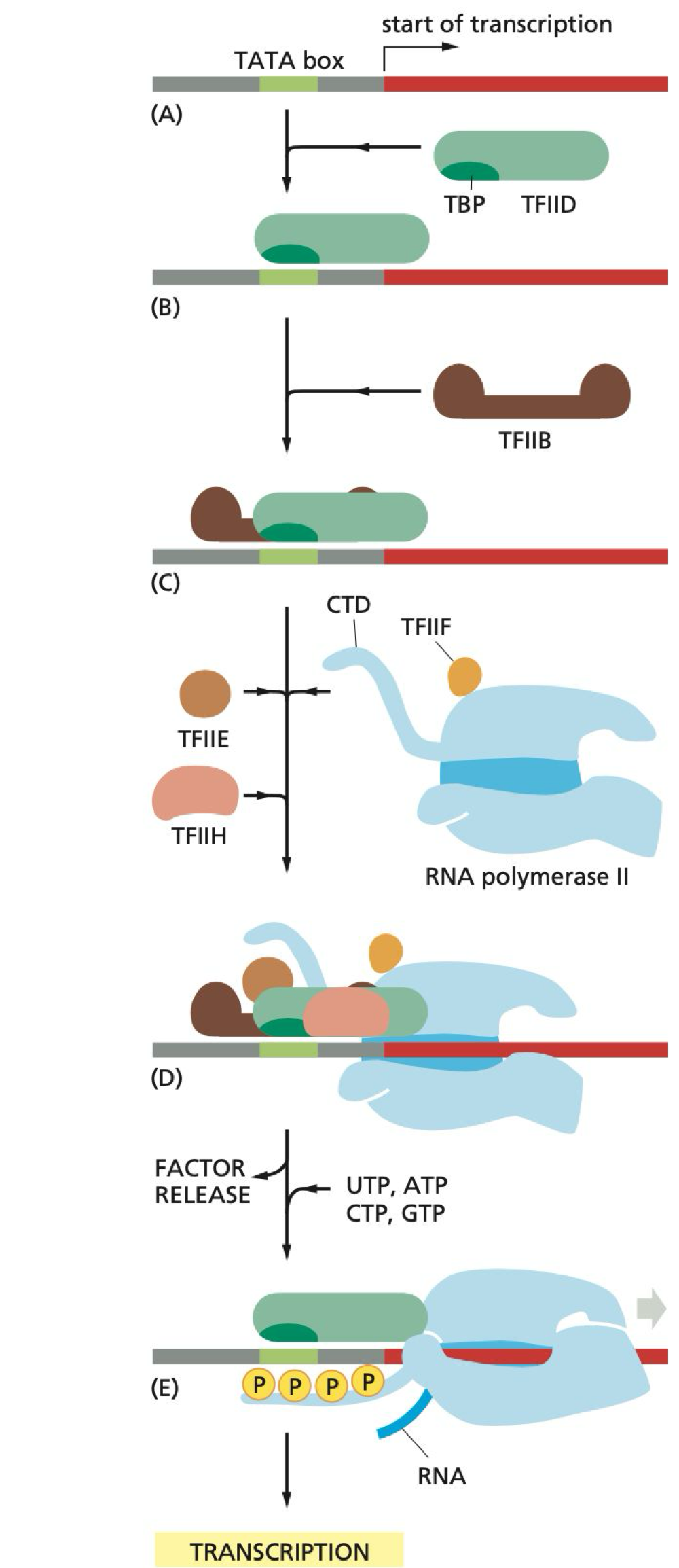
What happens after TFIIH phosphorylates Ser5?
This causes a conformational change and release of pol II from the promoter and transcription factors, allowing transcription to begin
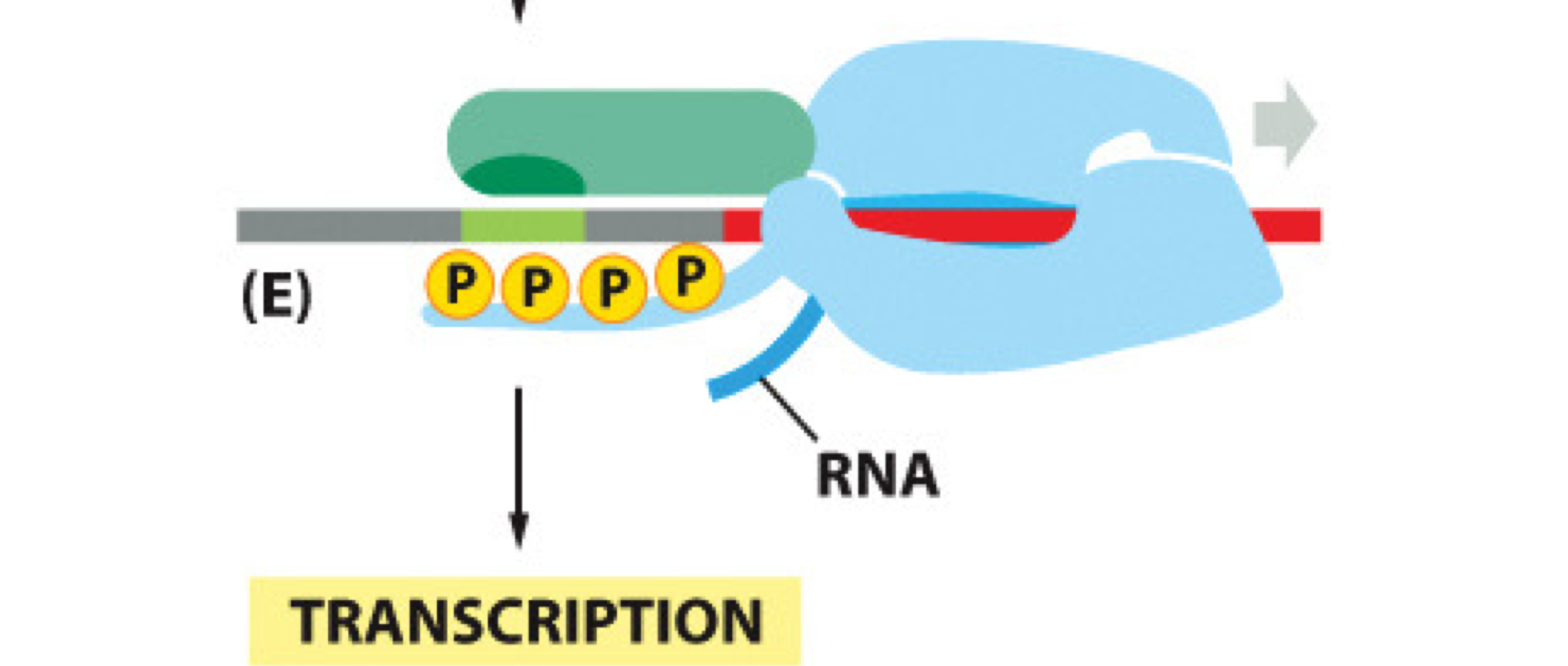
When is the 5' Guanine cap added to RNA?
The 5' Guanine cap is added shortly after transcription begins
What happens during the early elongation phase?
Pol II synthesizes 30-50 bp then pauses
What phosphorylation event allows pol II to continue elongation?
Ser2 on the CTD of pol II is phosphorylated, causing a conformational change that allows pol II to continue elongation
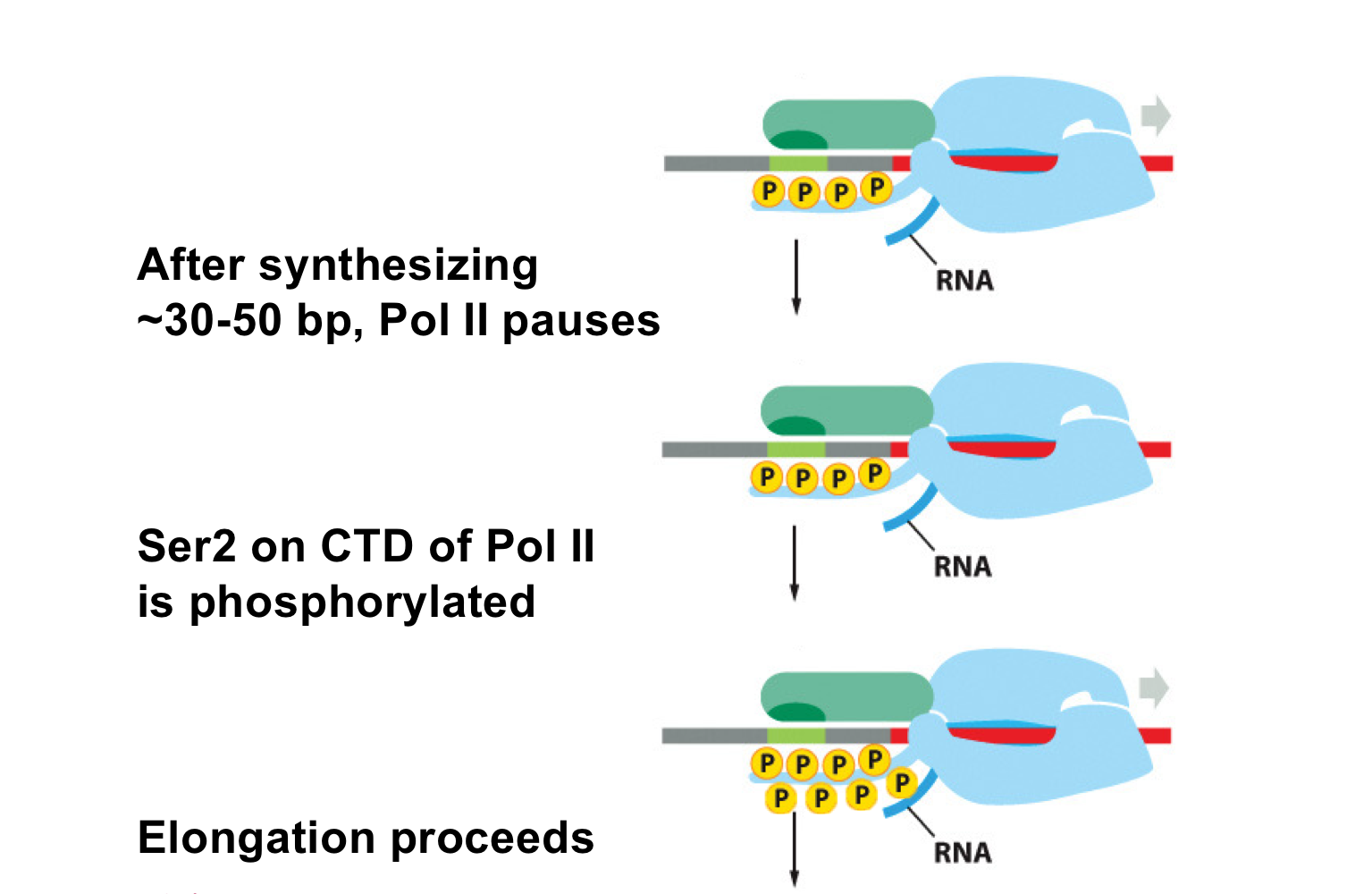
During elongation, which serines are phosphorylated on the CTD?
Both Ser5 and Ser2 are phosphorylated during elongation
What is the termination sequence for RNA pol II?
AAUAAA (on RNA) or TTATTT (on DNA template)
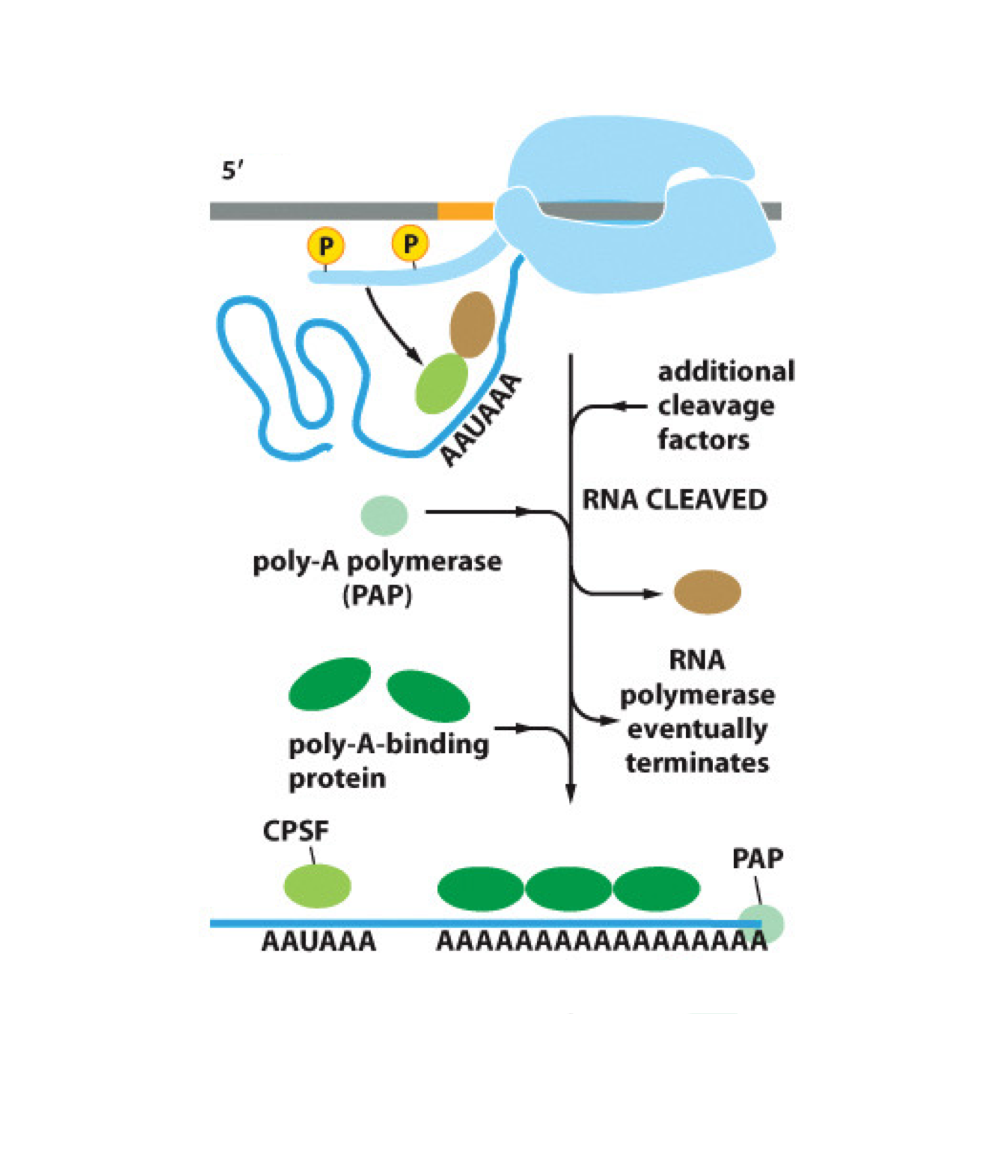
What is the "Torpedo model" of termination?
CstF and CPSF proteins transfer from pol II to bind the termination sequence, cleave and release the RNA transcript, while an exonuclease "chases" pol II and forces it to release
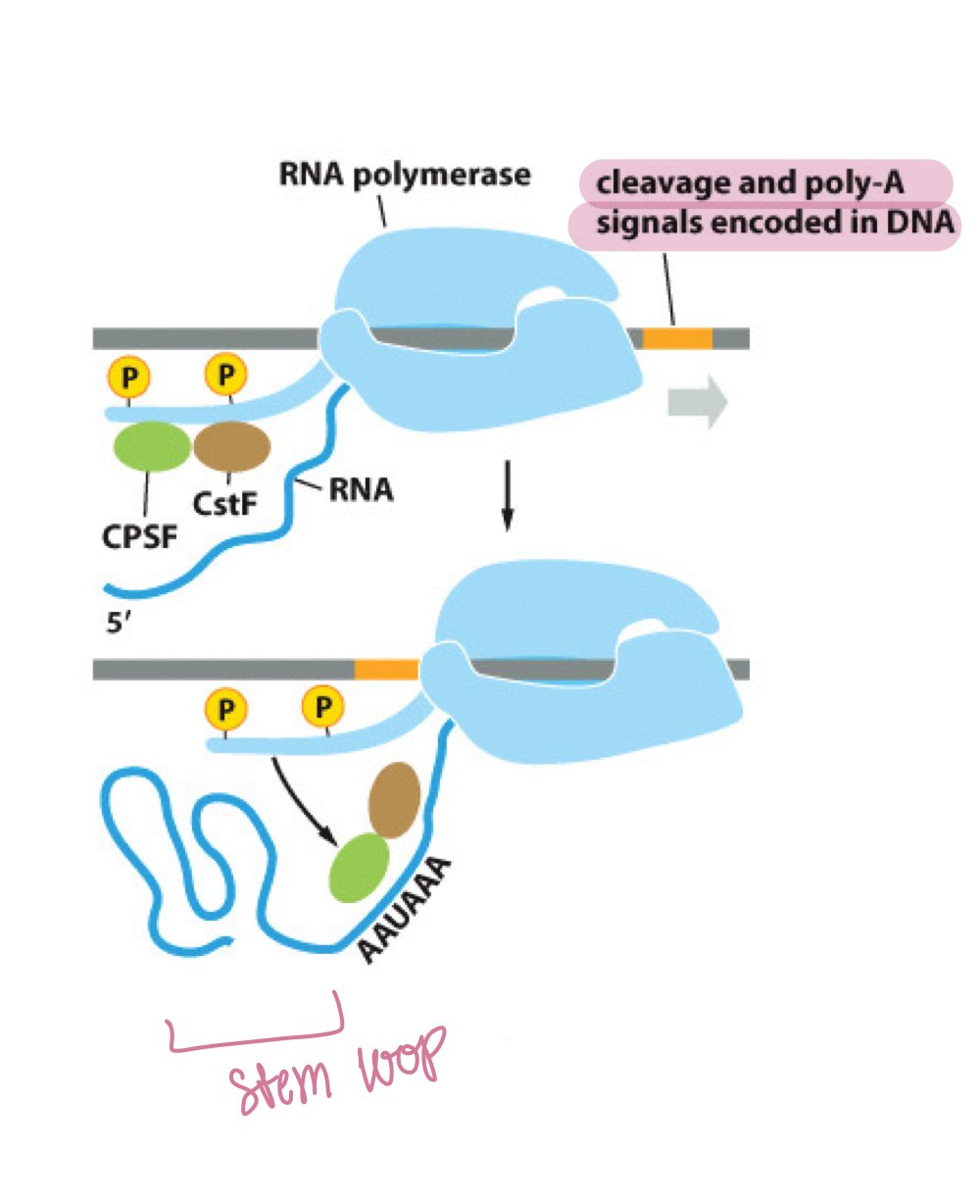
What proteins are involved in the torpedo model of termination?
CstF and CPSF proteins
What do CstF and CPSF do during termination?
They form hairpins and stem loops in RNA, weakening its association with pol II, and cleave the RNA transcript

What enzyme adds the poly-A tail?
Poly-A polymerase (PAP) adds the poly-A tail to the 3' end of RNA
What is the "Allosteric model" of termination?
The termination sequence causes a conformational change in pol II leading to spontaneous dissociation from the template strand
What phosphorylation change may occur during termination?
Some Ser5 in the CTD may be dephosphorylated
What is positive supercoiling?
DNA is overwound
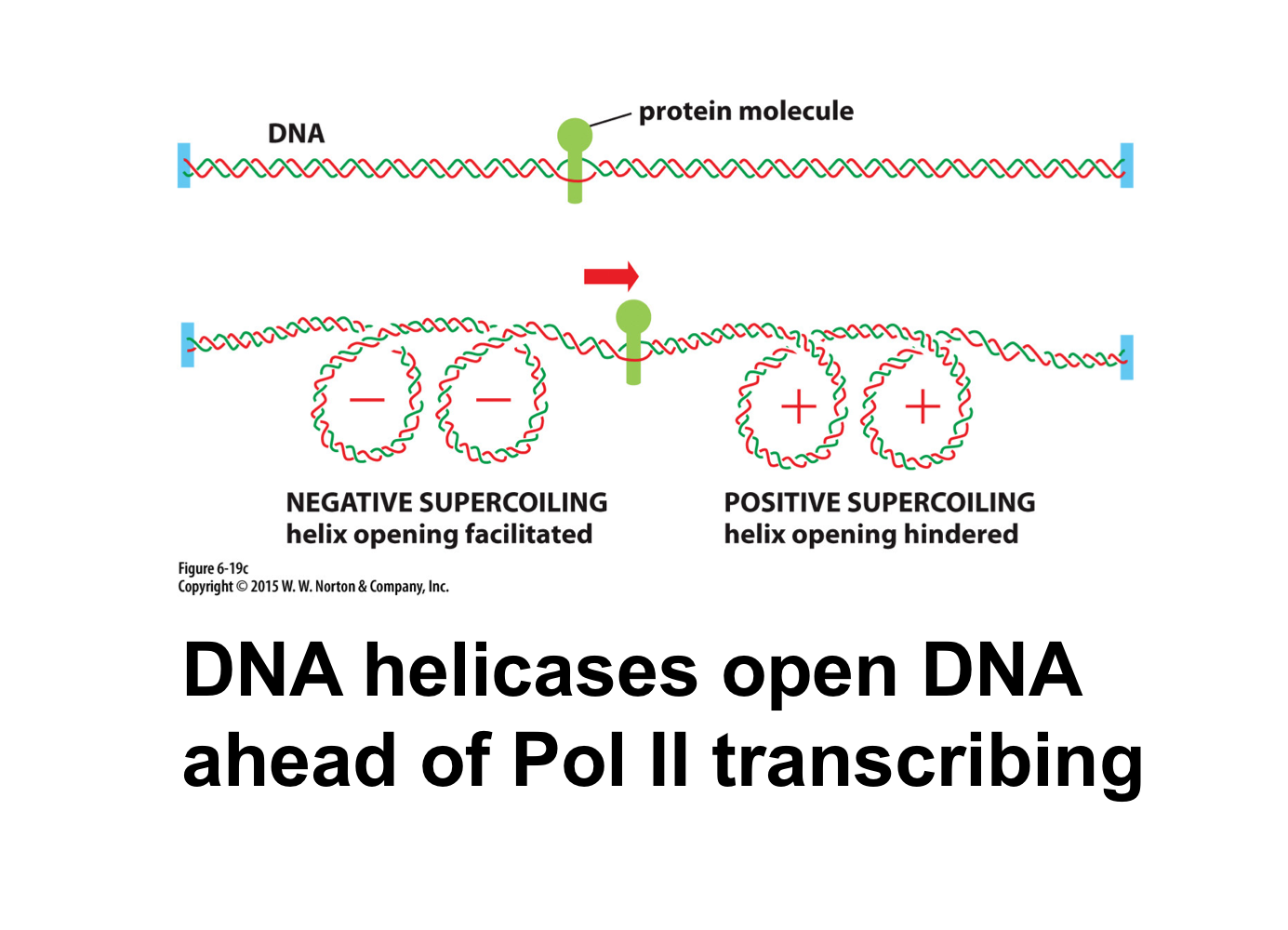
What is negative supercoiling?
DNA is underwound
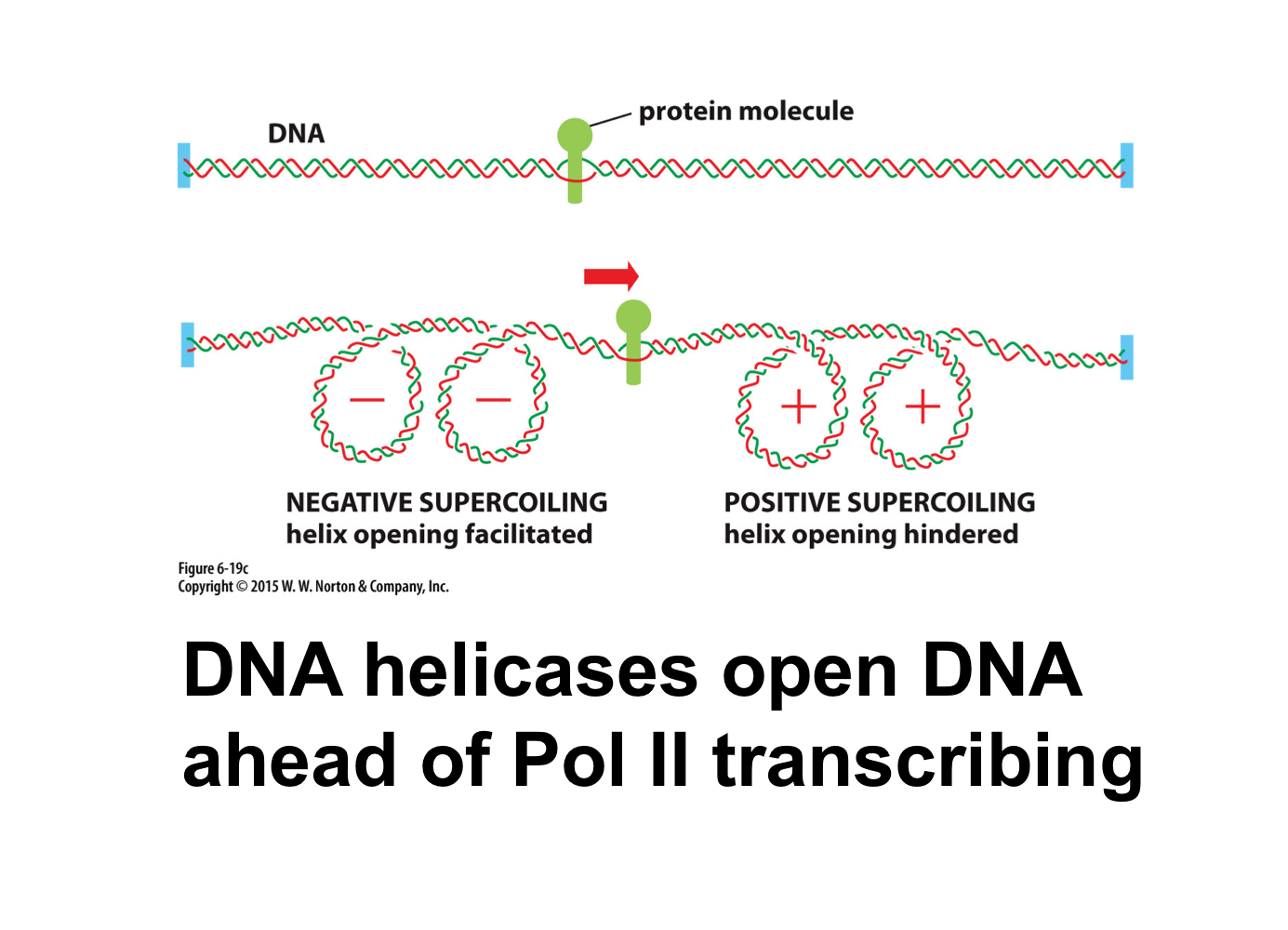
How often does the DNA helix rotate?
The helix rotates every 10 bp
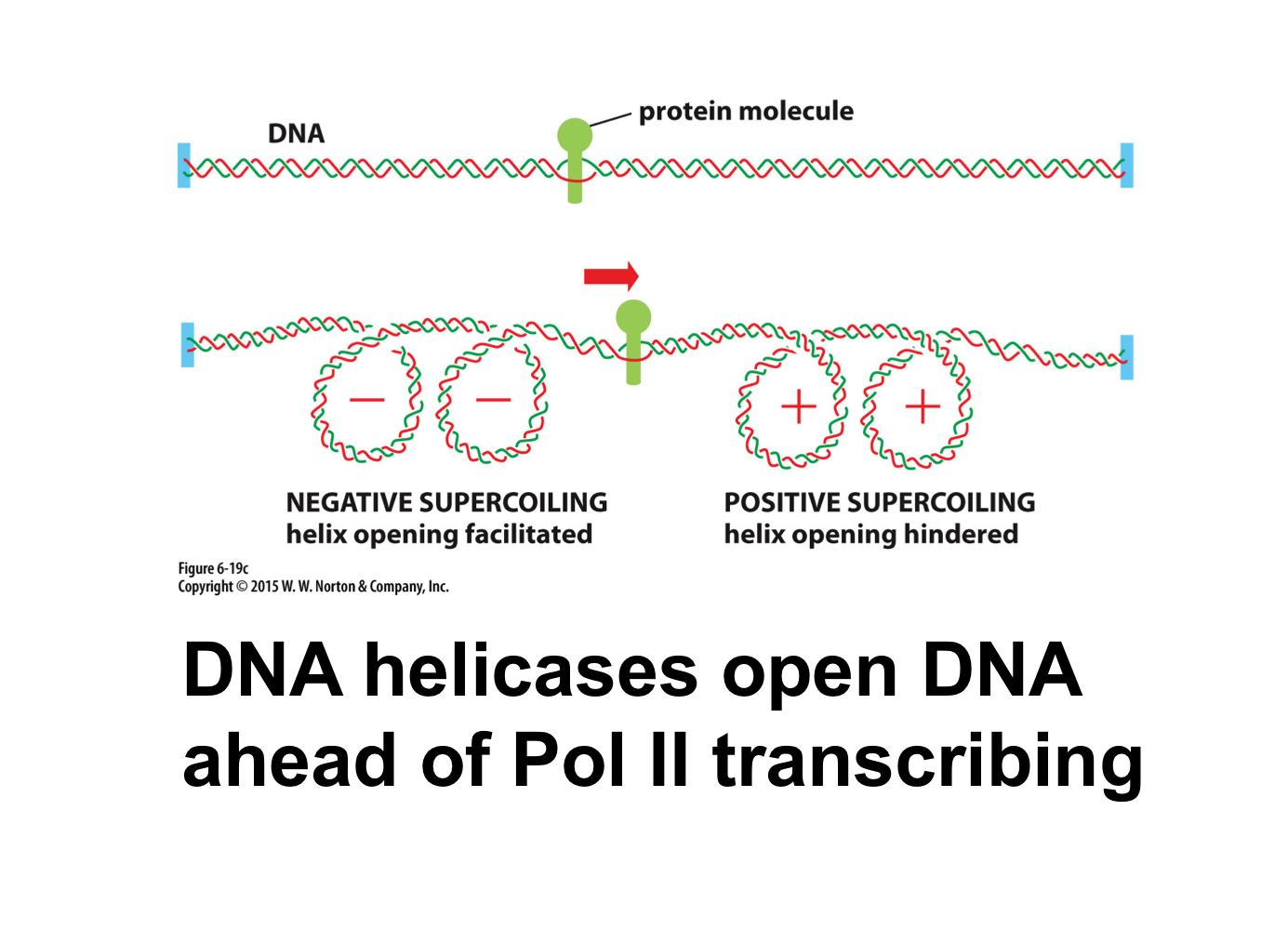
What supercoiling pattern occurs during transcription?
DNA helicase creates positive supercoiling ahead of transcription and negative supercoiling behind transcription
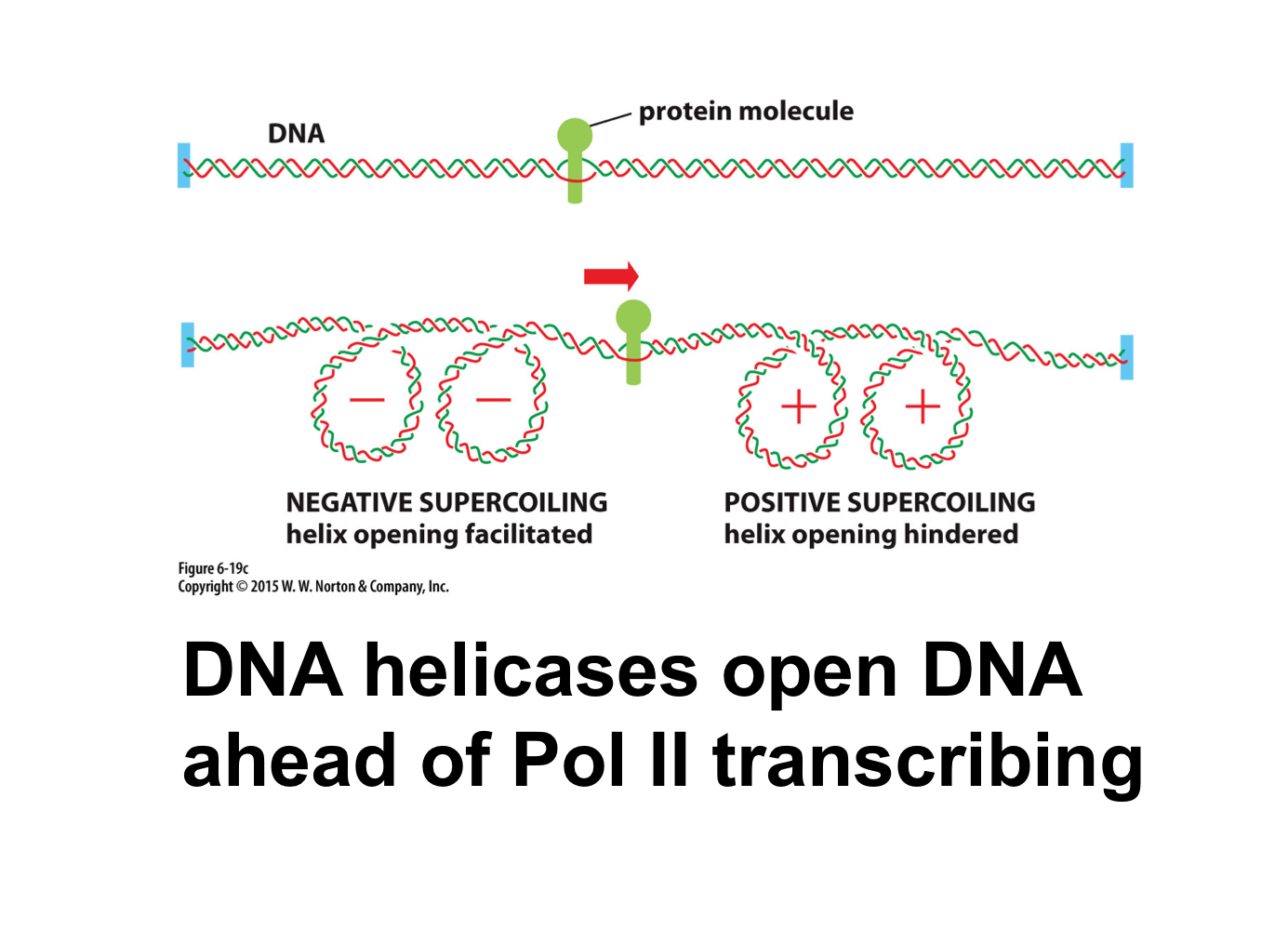
What are general transcription factors also called?
General TFs are also called basal TFs
Where do general transcription factors bind?
General TFs (TFIIB, TFIIH, TIIHD) bind to promoters
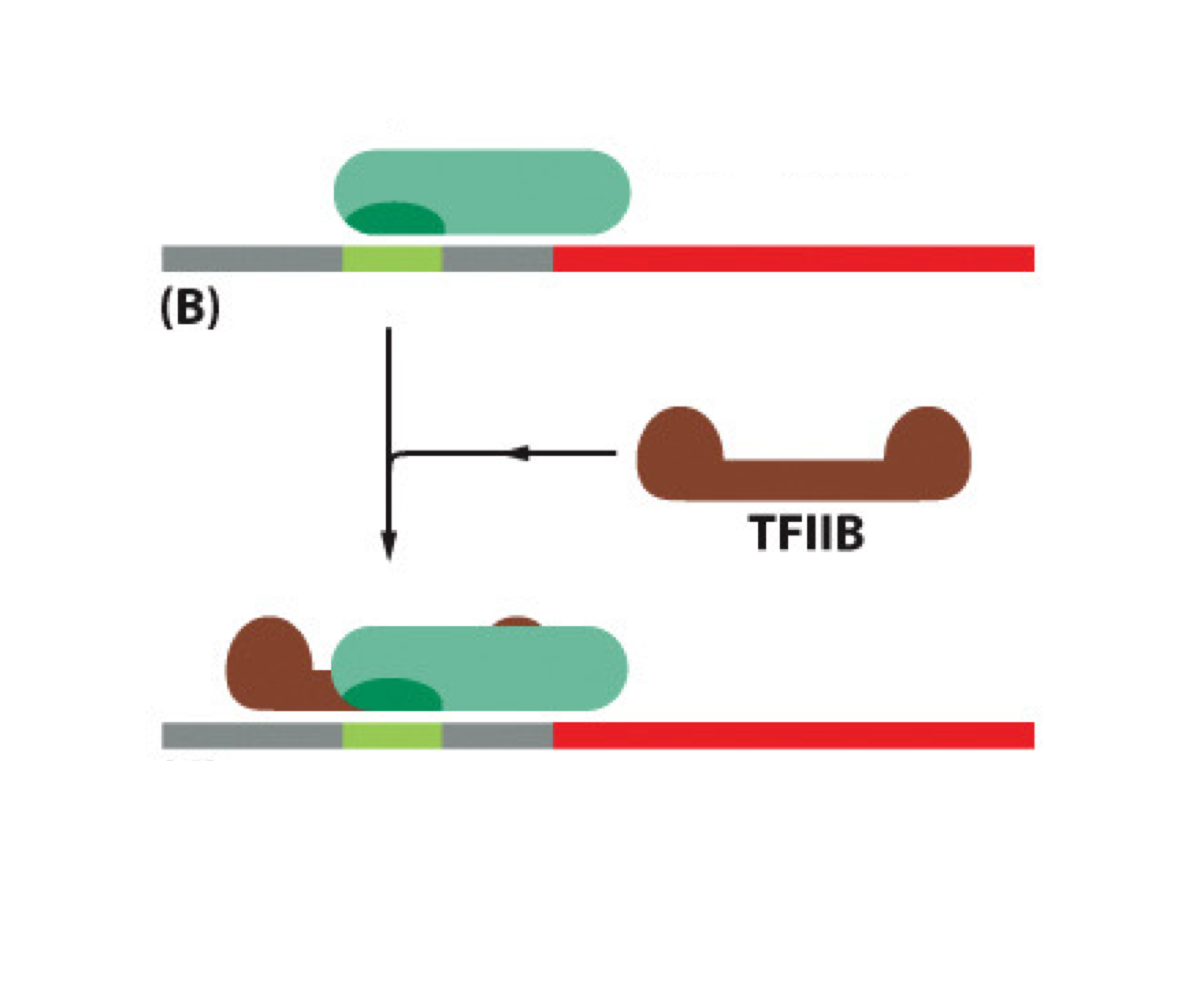
Where do sequence-specific transcription factors bind?
Sequence-specific TFs bind to enhancers (specific DNA sequences)
Do cofactors bind directly to DNA?
No, cofactors do NOT bind DNA directly. They bind to other TFs and bridge sequence-specific TFs and pol II
What makes pioneer transcription factors special?
Pioneer TFs can bind even when DNA is wrapped around nucleosomes and help open chromatin
What is the pre-initiation complex composed of?
General TFs + Pol II make up the pre-initiation complex at the promoter
What are enhancers?
DNA sequences that contain clusters of binding sites for sequence-specific TFs and increase transcription at promoters
What does it mean that enhancers are "modular"?
Modular means enhancers can work together with multiple other enhancers to control gene expression
What does it mean that enhancers are "autonomous"?
Autonomous means enhancers can work independently as well
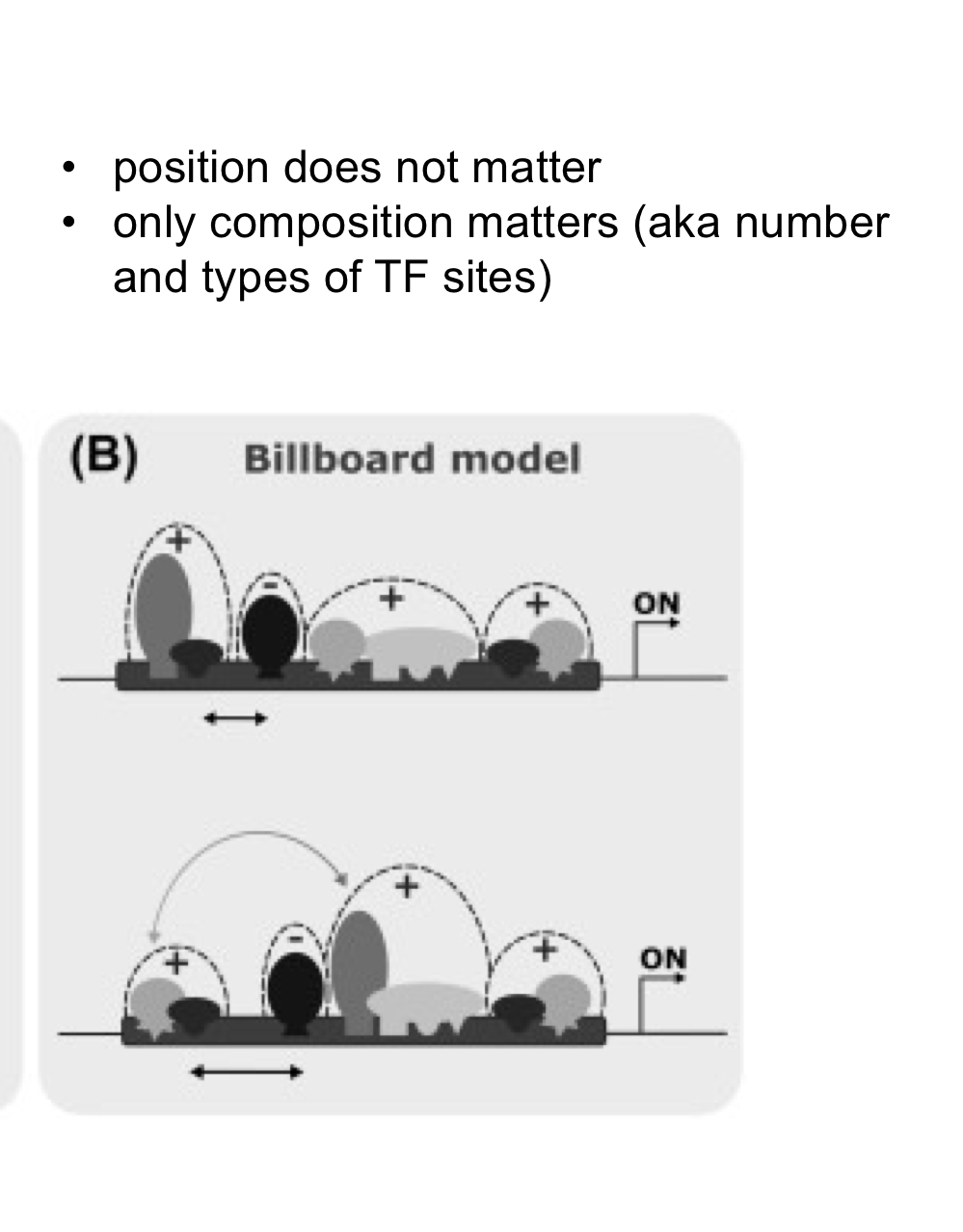
In the Billboard model of enhancer function, what matters?
In this model composition (number and types of binding sites) matters, but positioning does NOT matter. It's more flexible.
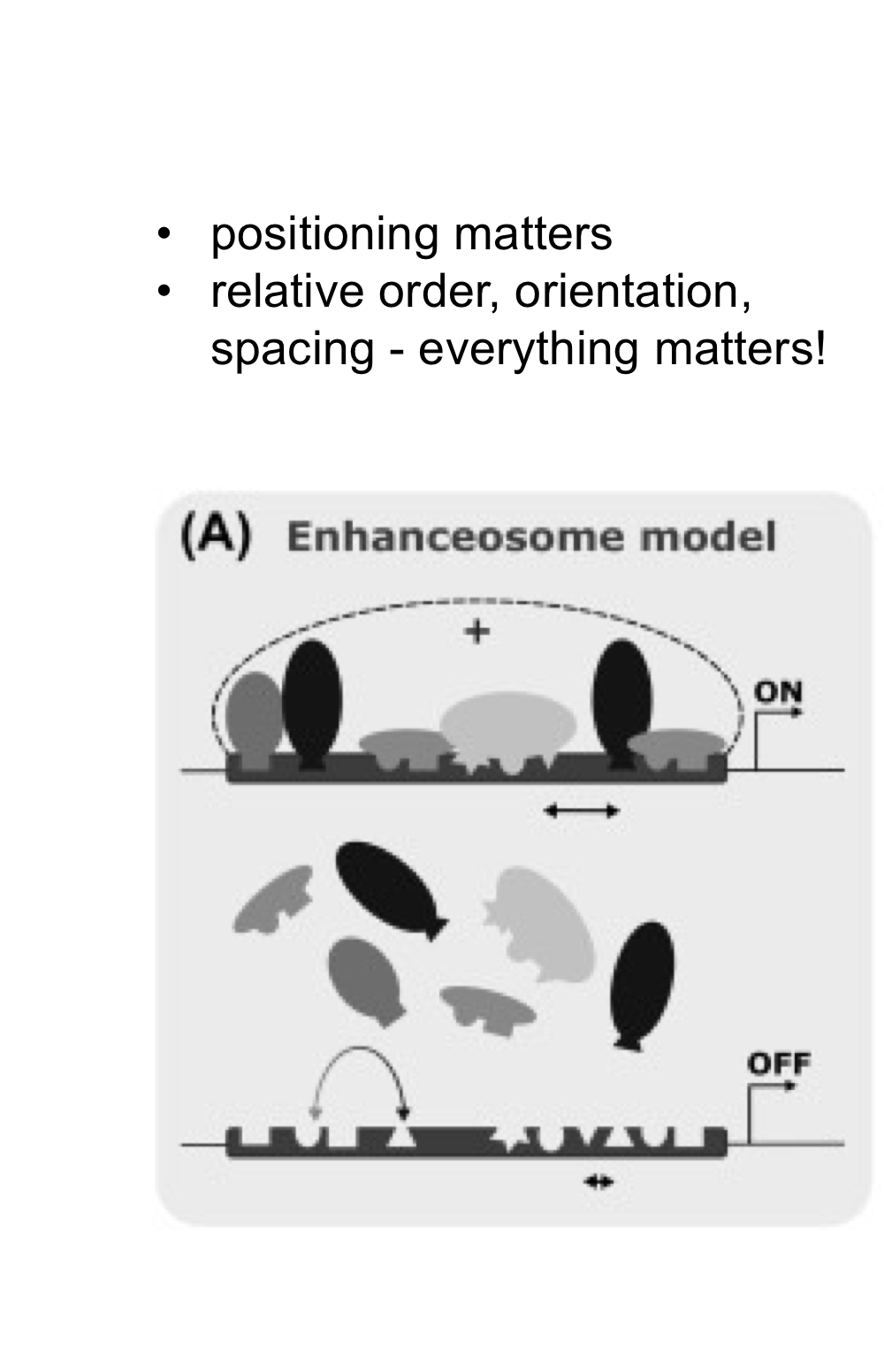
In the Enhanceosome model, what matters?
In this model, EVERYTHING matters - positioning, order, orientation, and spacing all matter. It's cooperative and highly specific.
How do nucleosomes affect TF binding?
Nucleosomes block TF binding to DNA because DNA is tightly coiled around histones, making binding sites inaccessible
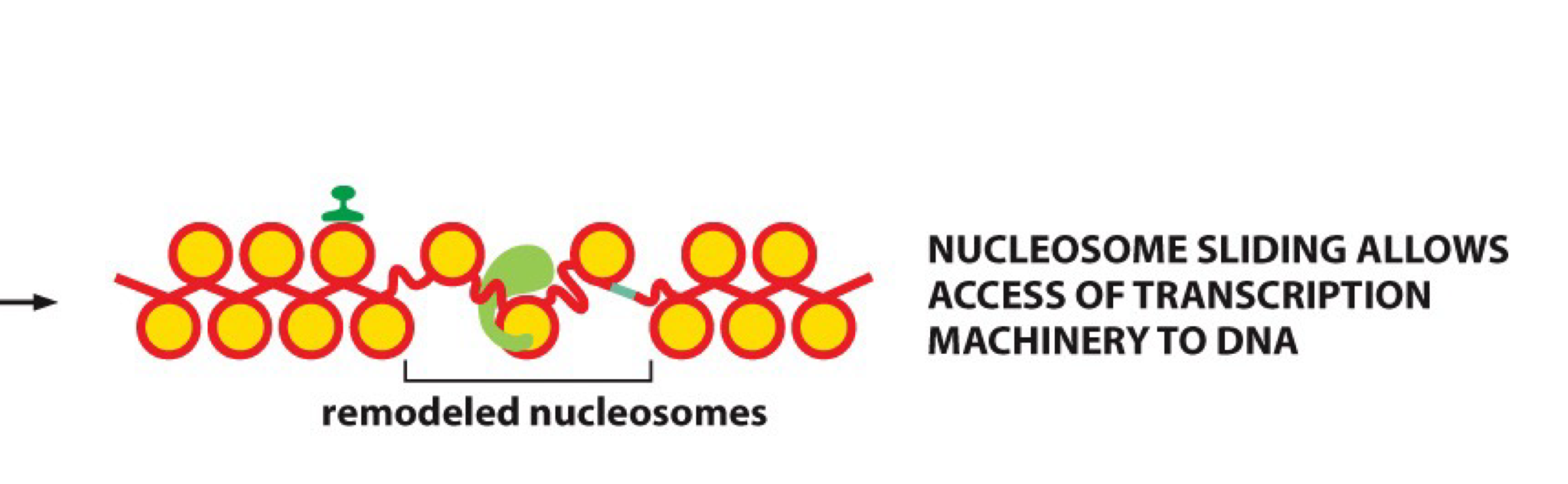
What are chromatin-remodeling complexes?
Complexes that move nucleosomes to expose binding sites for TFs
ATP driven process that causes slipping and sliding of histones
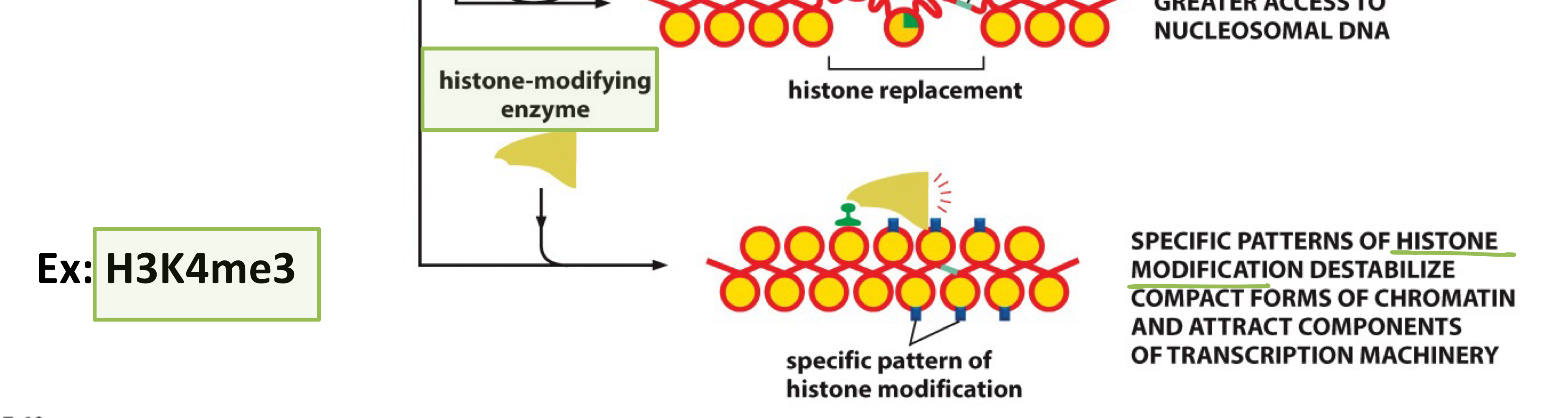
What do histone-modifying enzymes do?
TFs recruit these enzymes to add or remove chemical modifications (methylation, acetylation) to histone tails, weakening DNA-histone interactions
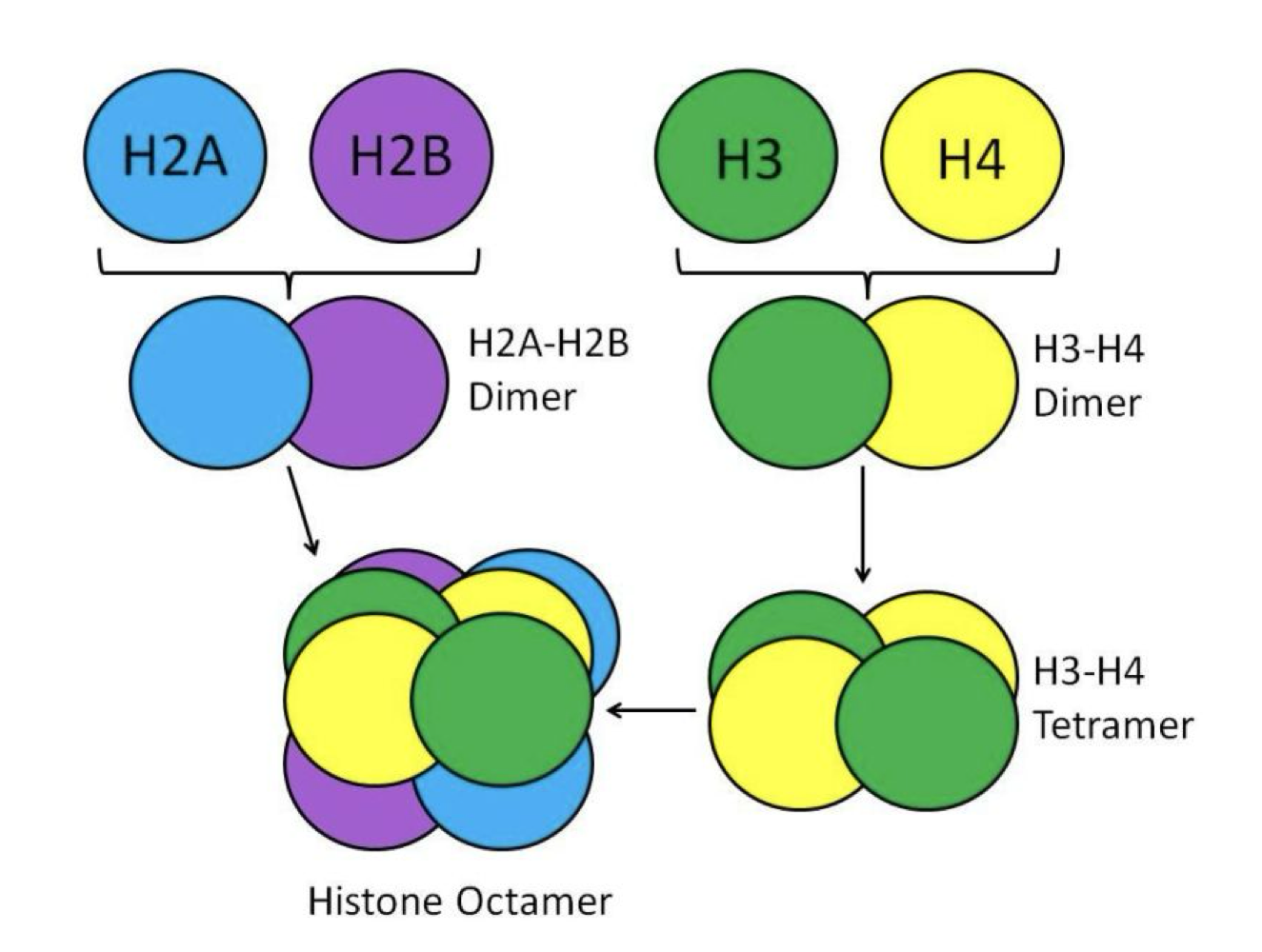
What is the composition of a nucleosome?
147 bp of DNA wrapped around a histone octamer 1.65 times. The octamer contains one H3/H4 heterotetramer (2 H3 + 2 H4) and two H2A/H2B heterodimers (2 H2A + 2 H2B)
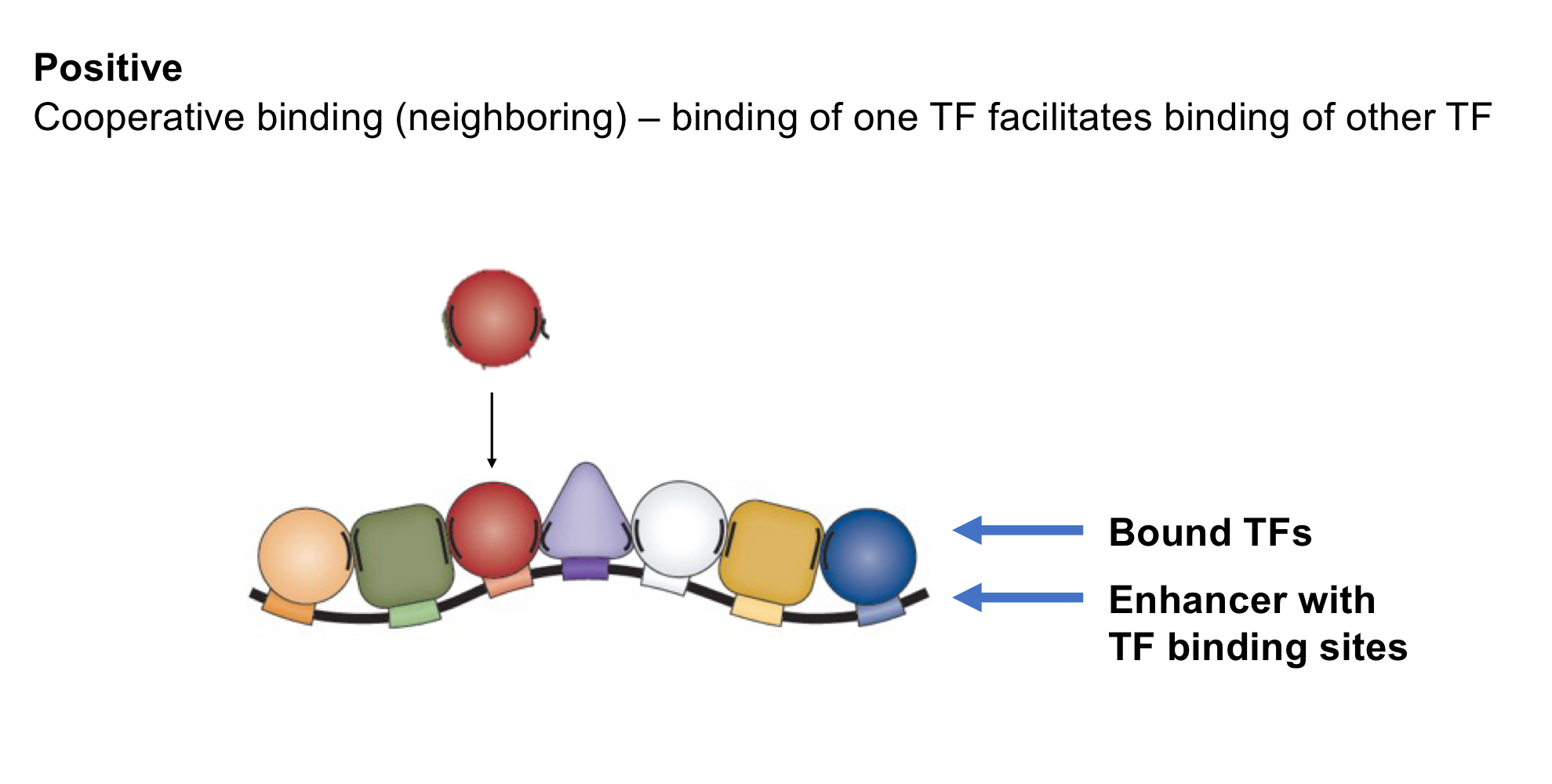
What is cooperative binding?
Positive TF interaction: When the binding of one TF to an enhancer makes the binding of another TF easier (increases binding affinity)
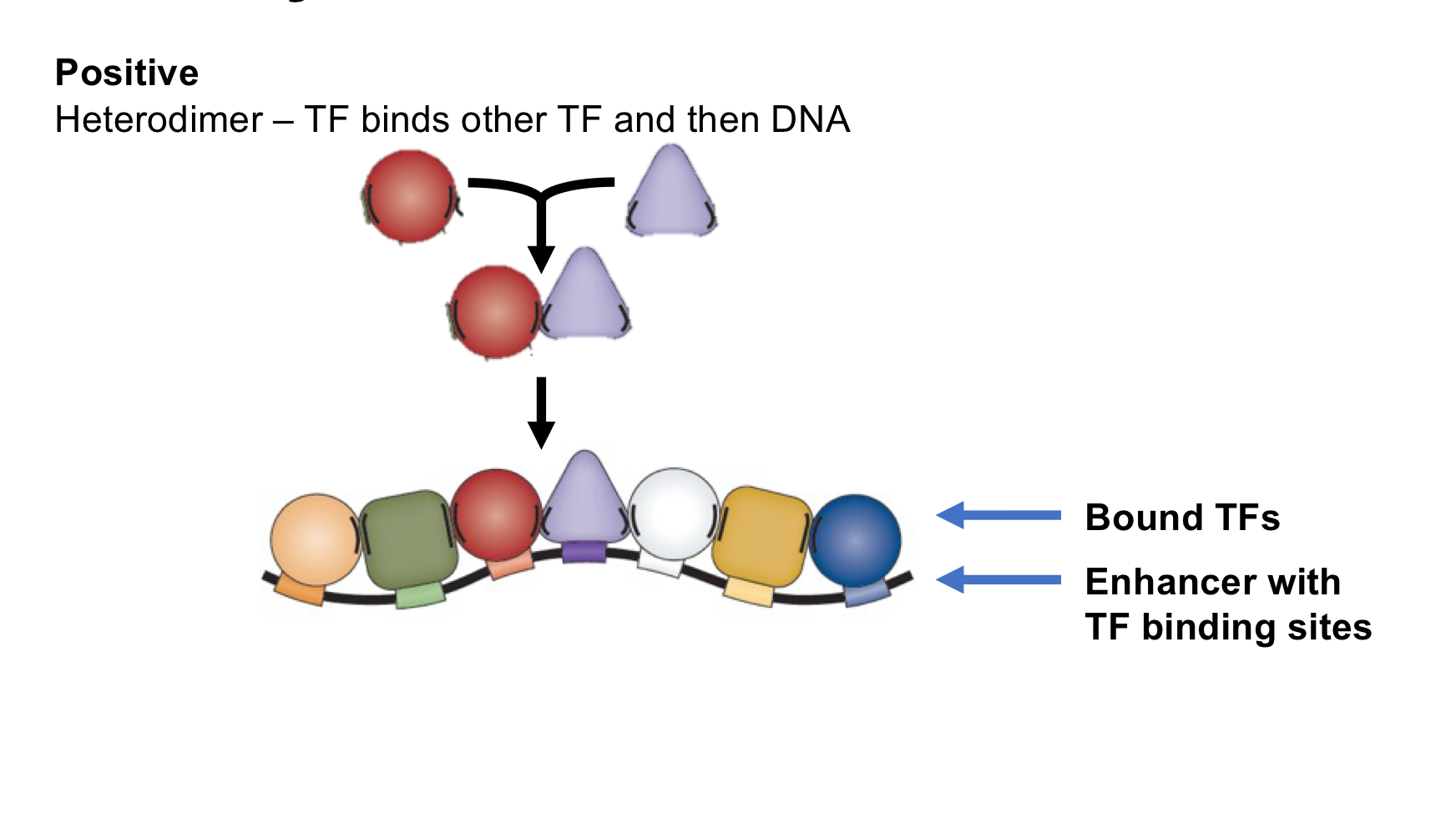
What is heterodimer formation?
Positive TF interaction: When two TFs bind to each other, then bind to the enhancer as a unit
What is co-occupancy?
Positive TF interaction: Rapid alternating (switching) where two TFs compete for the same binding site on an enhancer
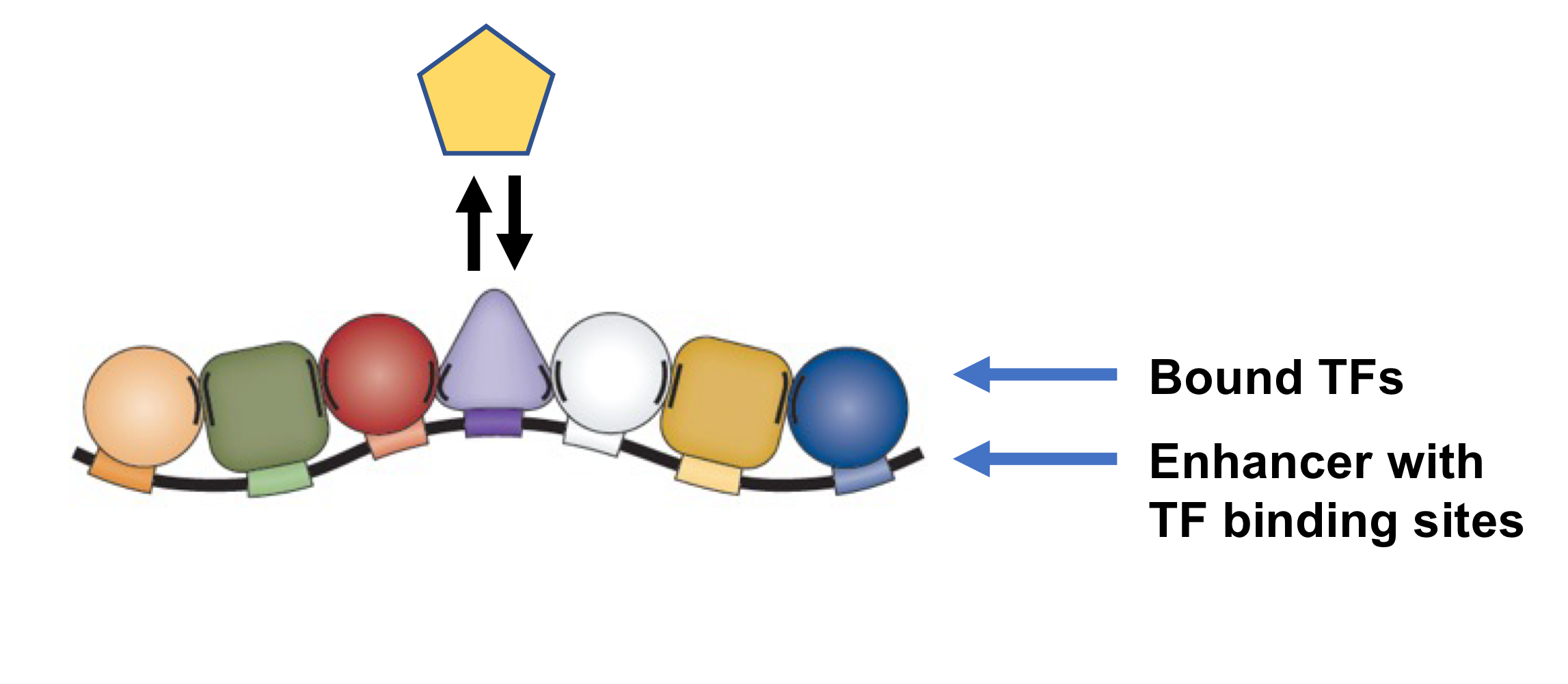
How do activators and repressors interact negatively? (Activator vs Repressor)
Activators (activate transcription) and repressors (inhibit transcription) compete for the same binding site on enhancers
What happens when a weaker activator replaces a stronger activator?
Negative TF interaction: Transcription decreases when a weaker activator replaces a strong activator in the binding site
What are the three steps of PCR?
1) Denature (use heat to separate DNA strands), 2) Anneal (primers stick to sequence-specific sites), 3) Elongation (synthesize new strand using DNA polymerase)
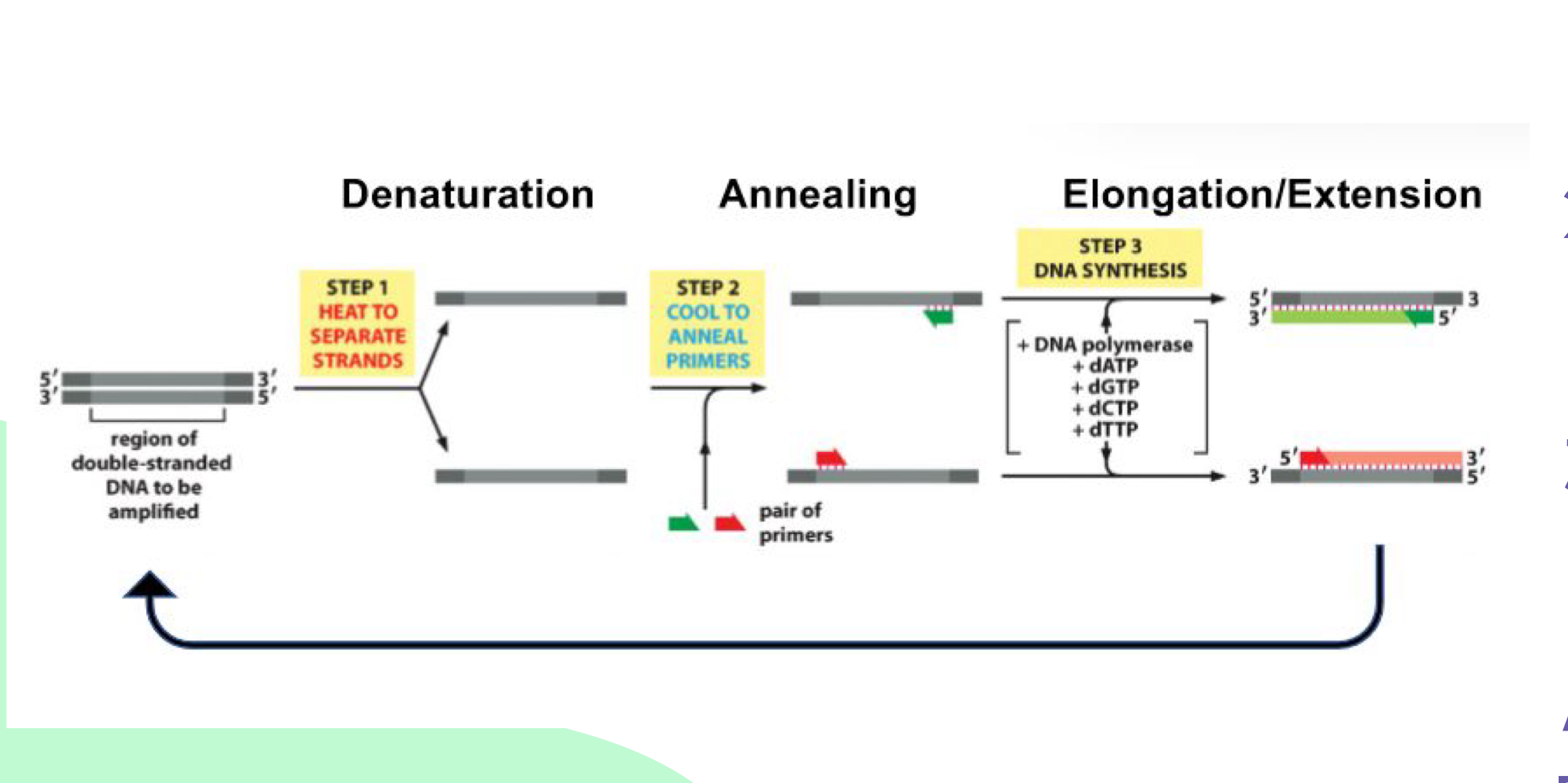
What is needed for RNA amplification in PCR?
Reverse transcriptase to create complementary DNA (cDNA) from RNA
How does gel electrophoresis separate DNA?
DNA fragments separate by size/molecular weight in an negative electric field. Small fragments move faster to the bottom, larger fragments move slowly and stay higher on the gel
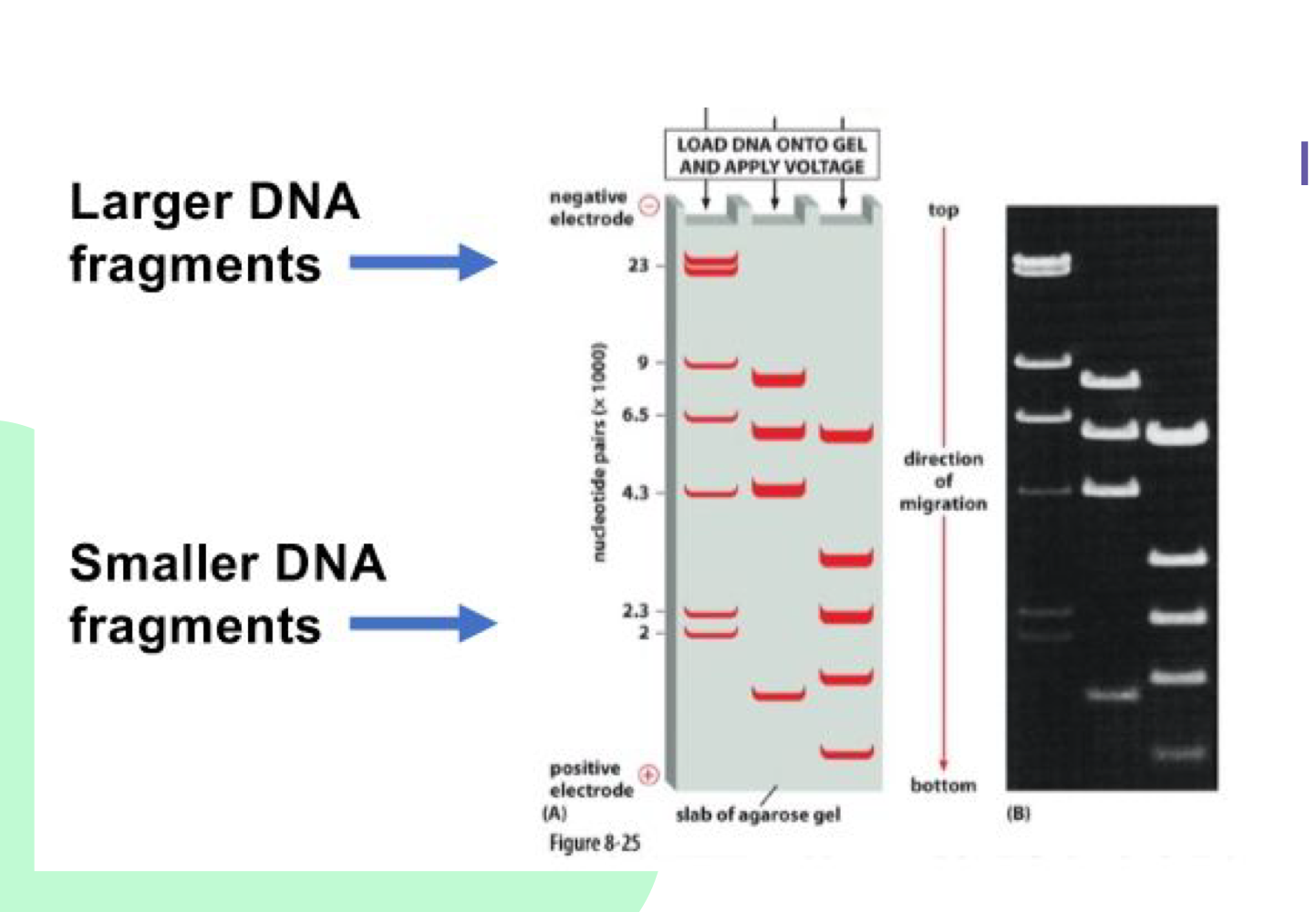
Why does DNA move toward the positive electrode?
Because DNA is negatively charged, it moves toward the positive electrode
What does a thick band indicate in gel electrophoresis?
A thick band indicates lots of product
What are adapters used for in Illumina sequencing?
Adapters are added to both ends of DNA fragments and contain sequences for binding to the flow cell and sequencing primers
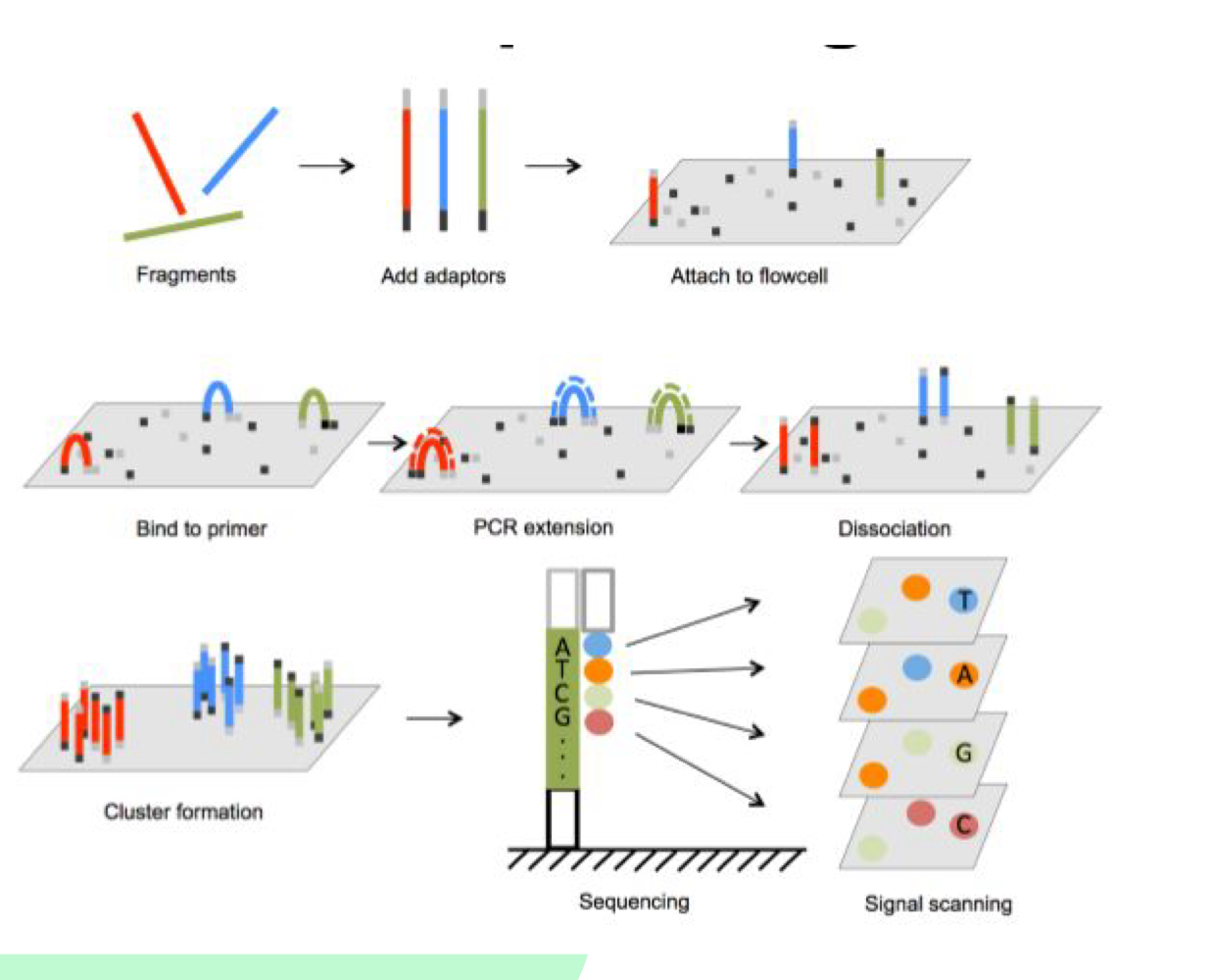
What happens during cluster generation?
Isothermal amplification occurs - fragments attach to flow cell, bind to complementary primers, DNA polymerase synthesizes new strands, creating bridge structures
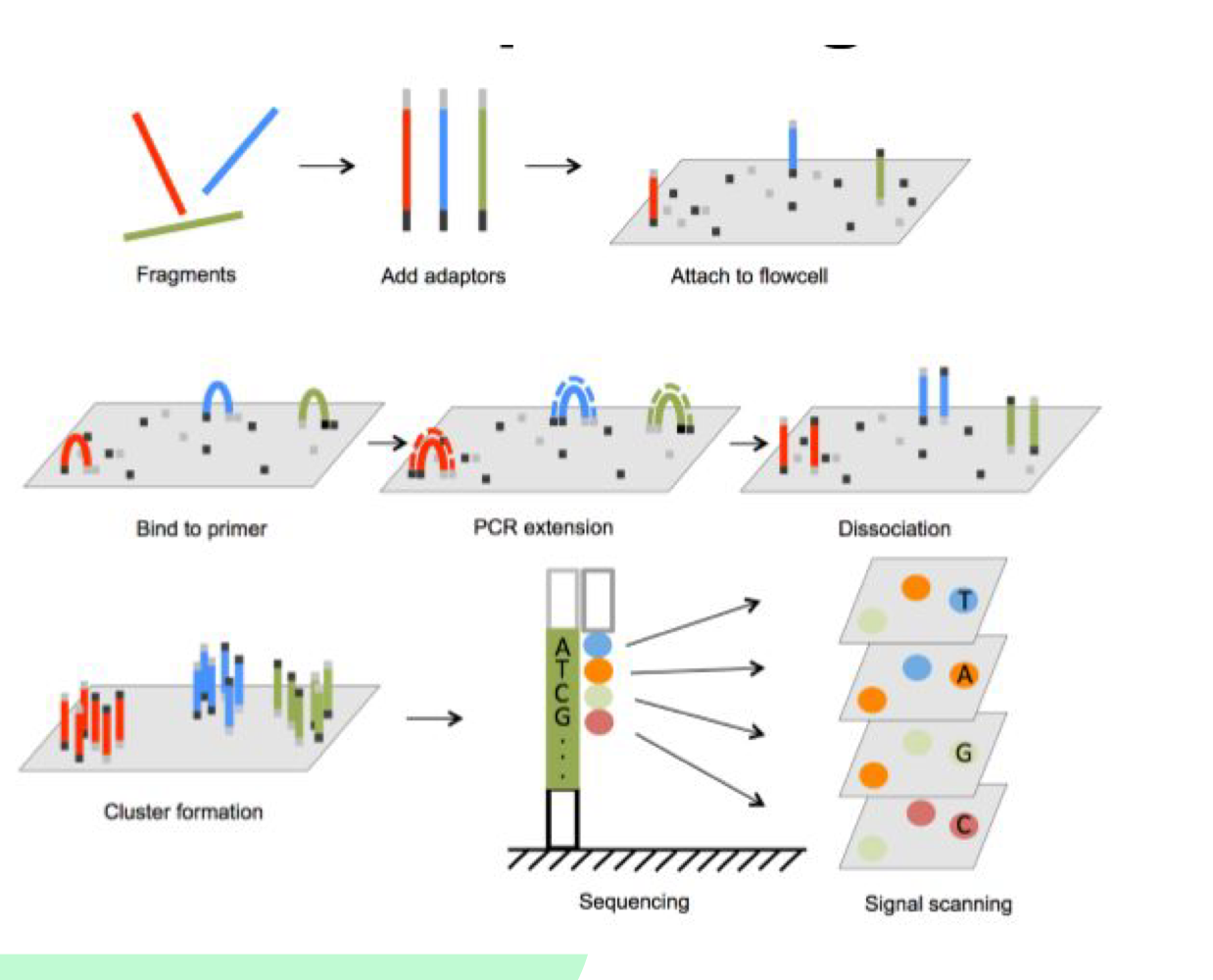
How does Illumina sequencing detect bases?
DNA polymerase adds nucleotides with fluorescence to the flow cell. Different colors correspond to each base (A, C, T, G)
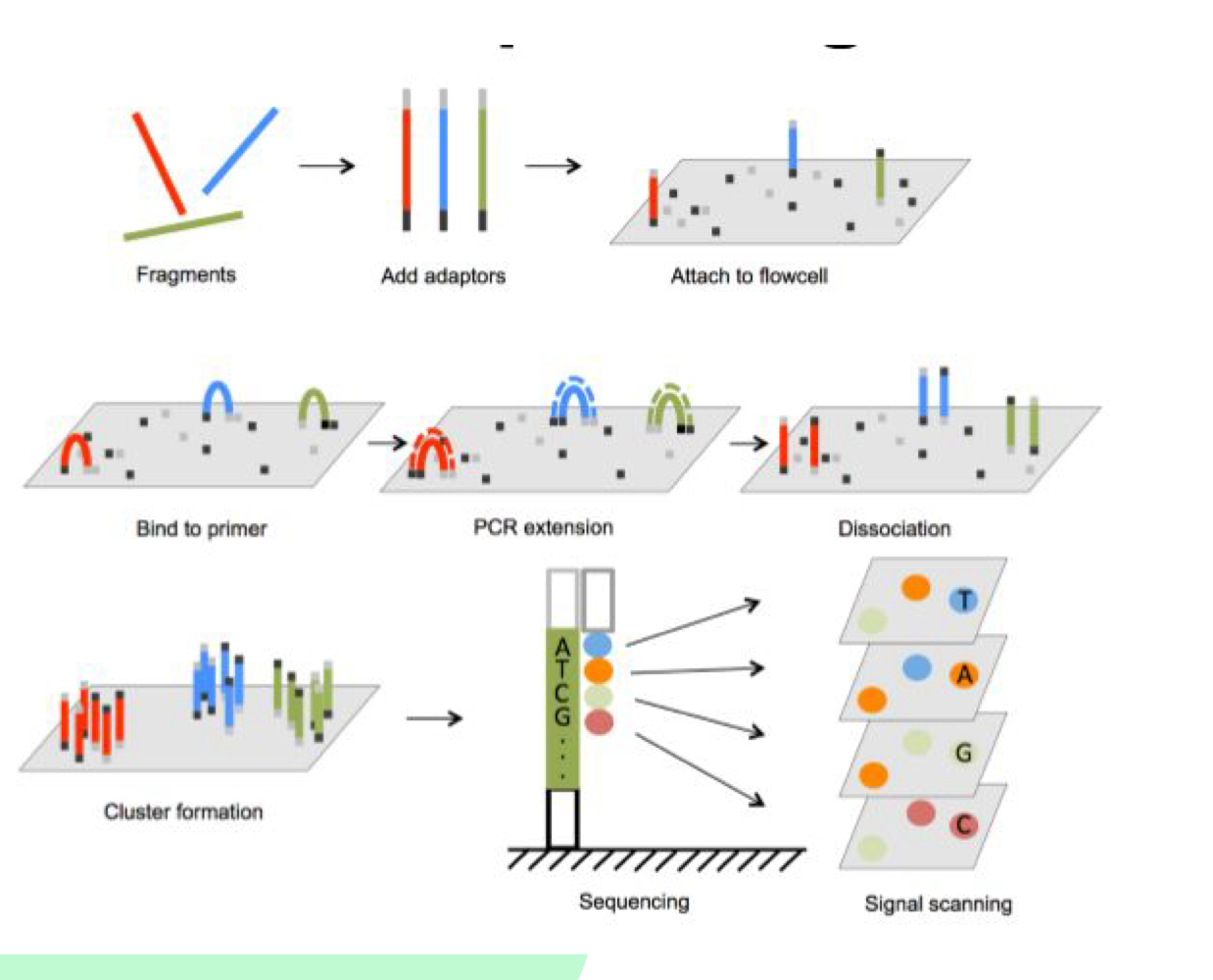
What happens during data analysis in Illumina sequencing?
Light flashes are recorded for each nucleotide, color and intensity translate to base sequences, and results are aligned with reference genome
What does RNA-Seq measure?
RNA-Seq measures gene expression levels
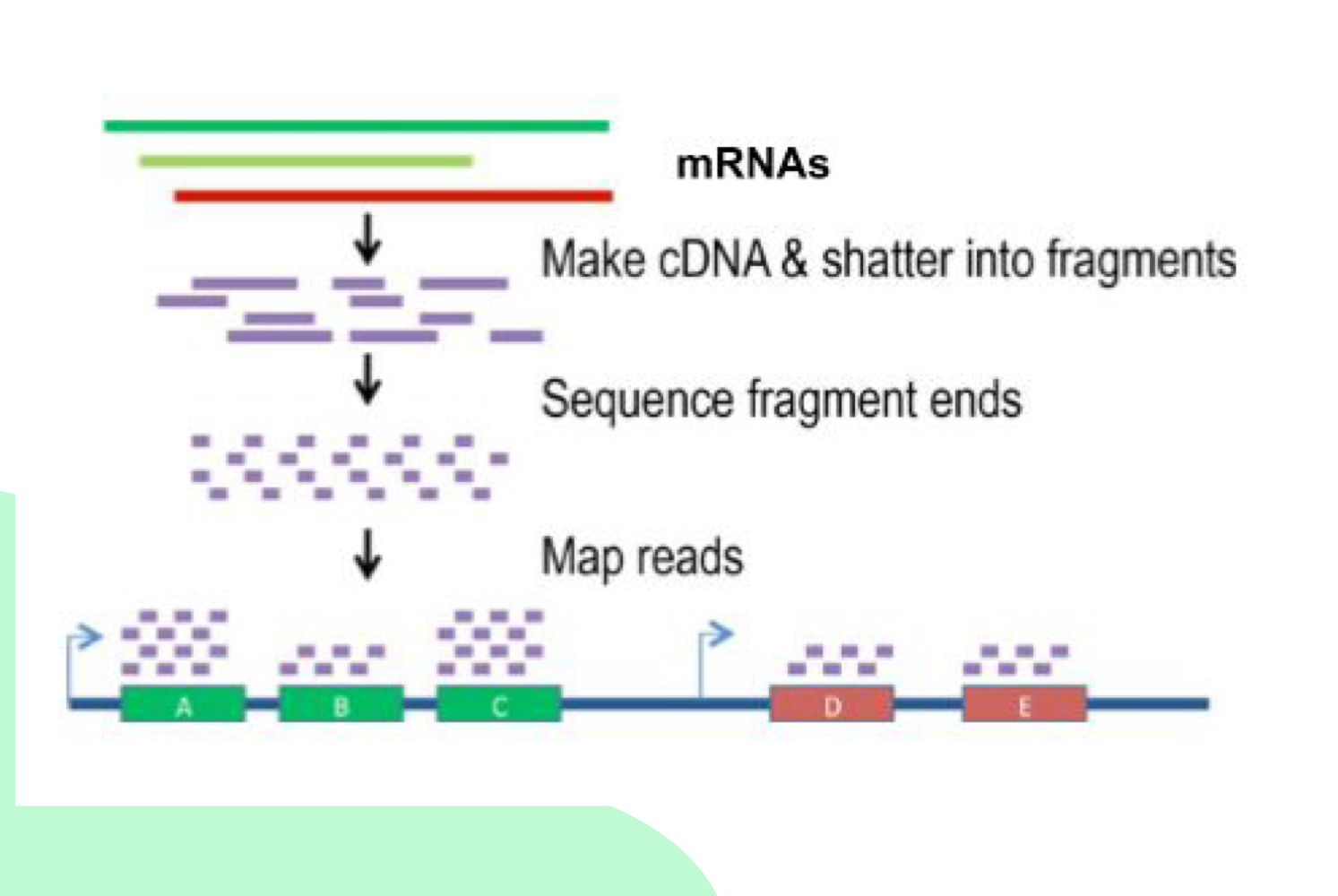
What are the main steps of RNA-Seq?
1) Get mRNA, 2) Use reverse transcriptase to make cDNA, 3) Shatter DNA into fragments, 4) Sequence fragment ends, 5) Map reads onto genome to look at expression levels
What does ChIP-Seq identify?
ChIP-Seq identifies specific DNA sequences bound to proteins (histones or transcription factors)
What is the difference between ChIP-qPCR and ChIP-Seq?
ChIP-qPCR looks at specific TF binding to a specific DNA sequence, while ChIP-Seq looks at specific TF binding to all bound DNA sequences in the genome
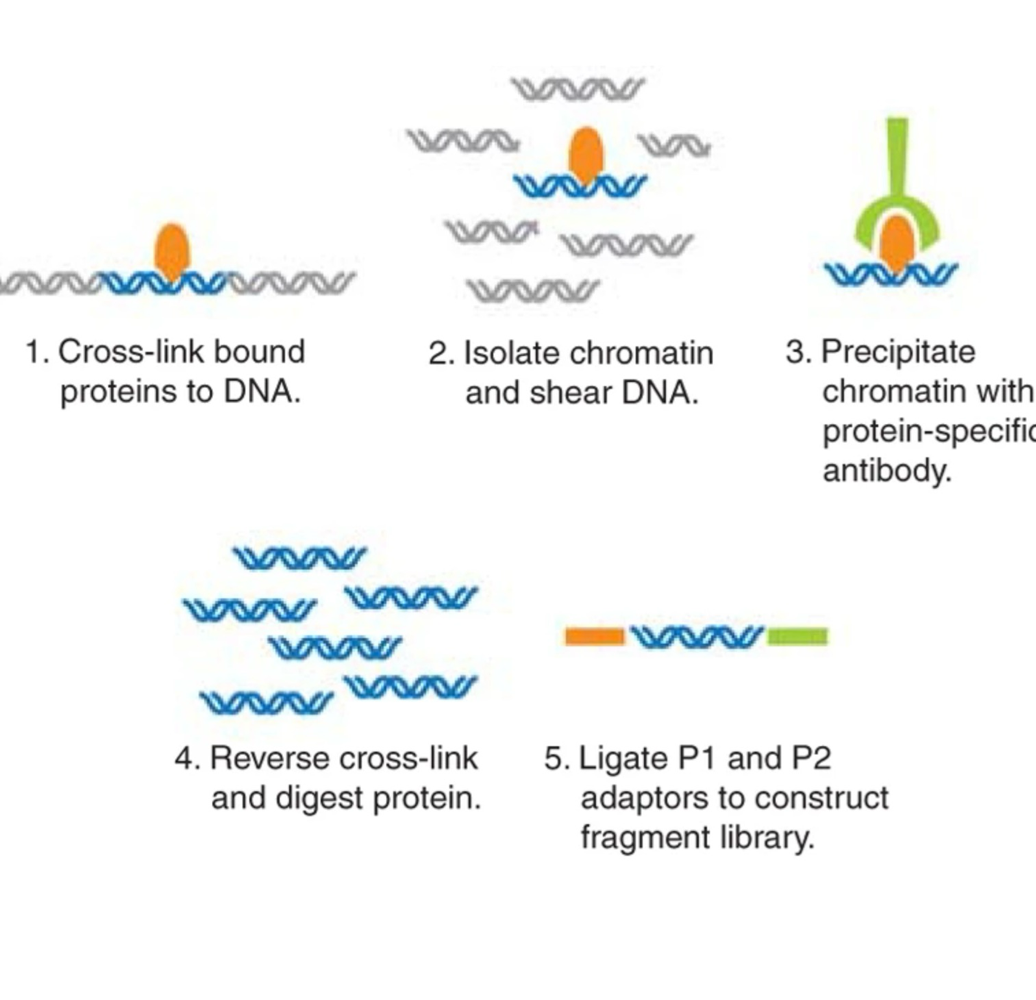
What happens during chromatin preparation in ChIP-Seq?
Proteins are cross-linked (glued) to DNA and DNA is broken into fragments
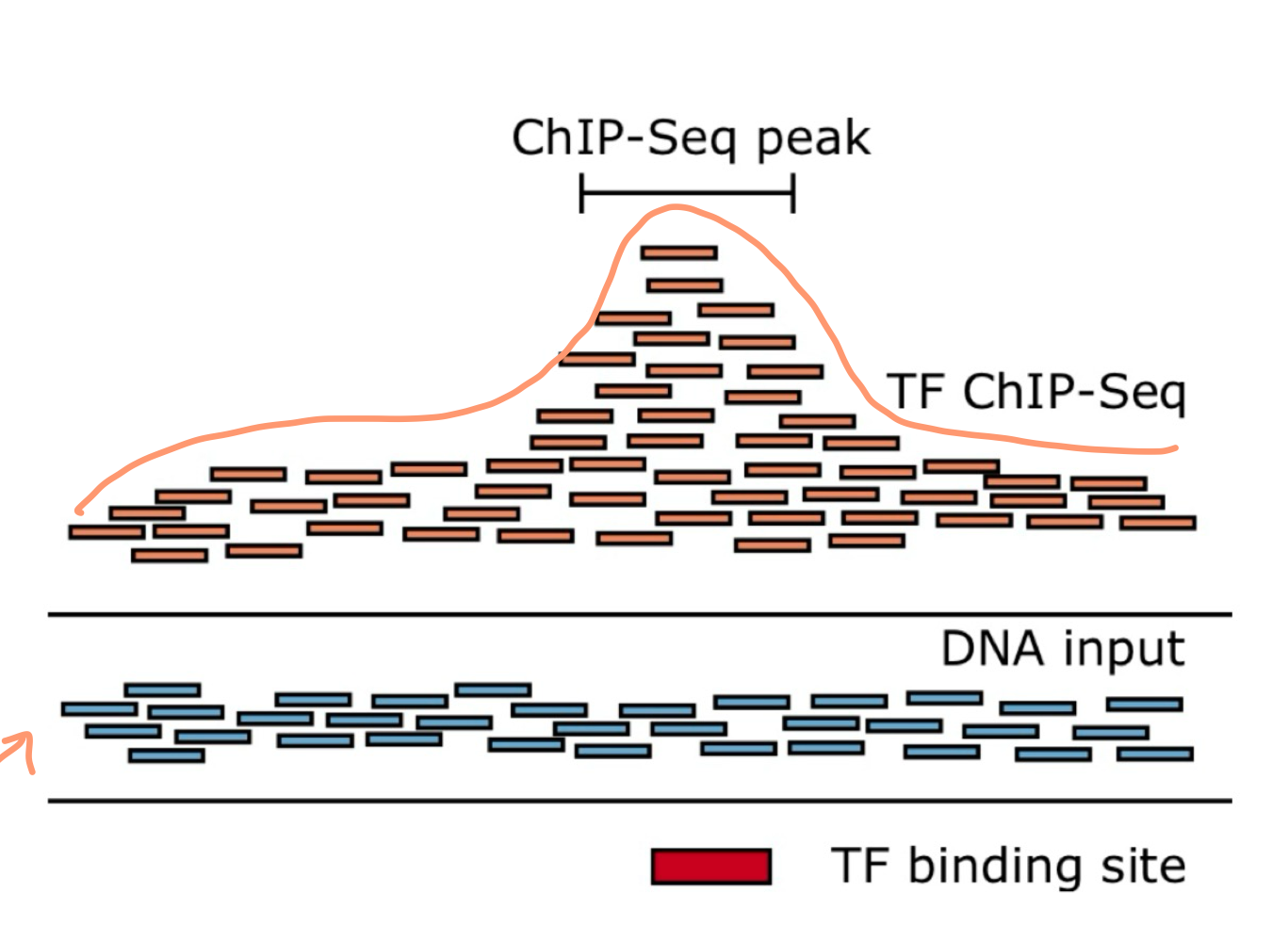
What do peaks represent in ChIP-Seq results?
Peaks represent where the protein was bound to DNA
What does bisulfite sequencing detect?
Bisulfite sequencing detects methylated (cytosine) DNA
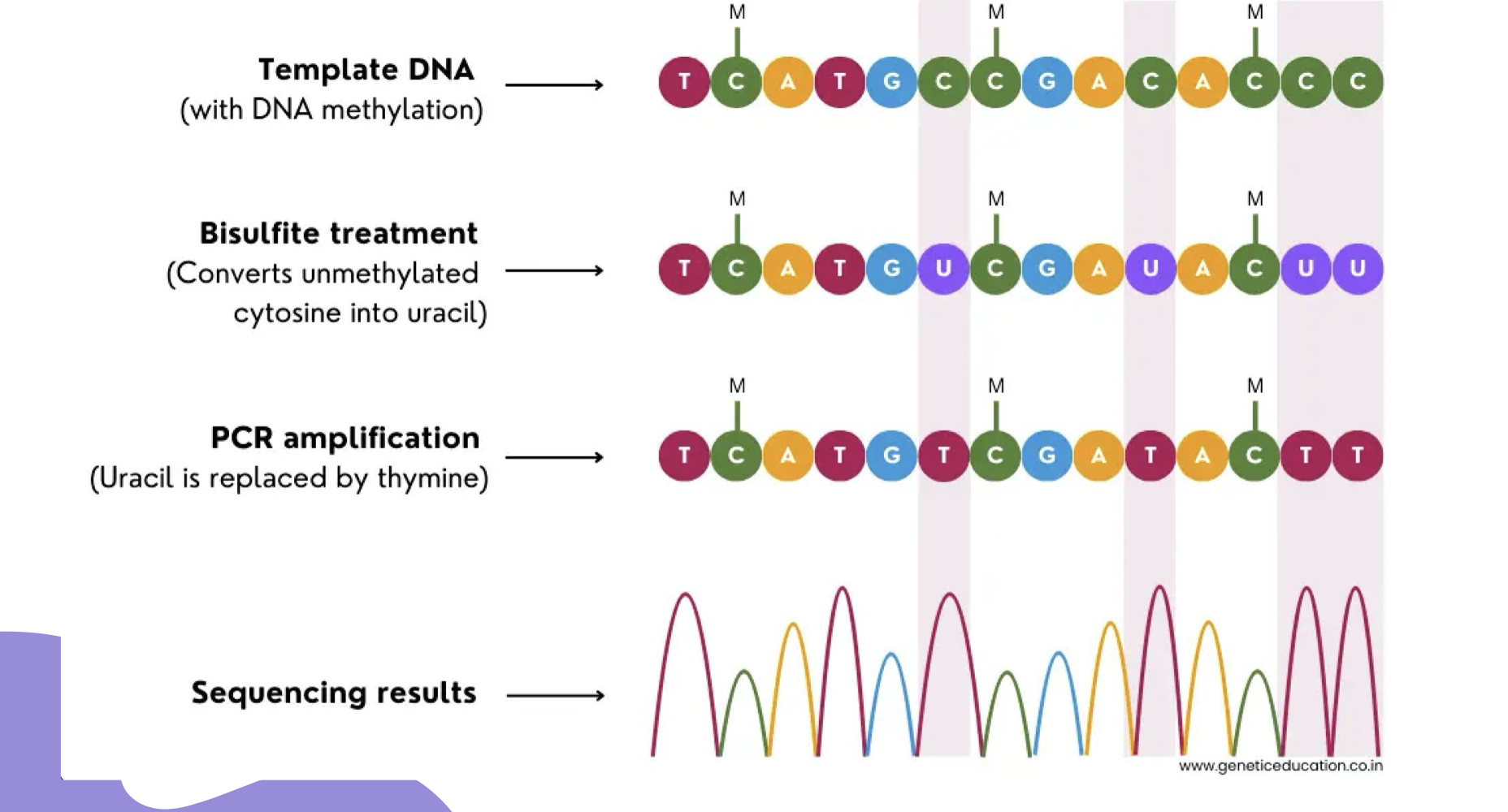
What happens during bisulfite conversion?
Unmethylated cytosine is converted to uracil, leaving methylated cytosine unaffected. Eventually the uracil is converted to thymine
What happens to uracils during PCR amplification in bisulfite sequencing?
The uracils are eventually converted to thymines

How do you interpret bisulfite sequencing results?
All remaining cytosines in the sequence are methylated (unmethylated ones were converted to thymine)
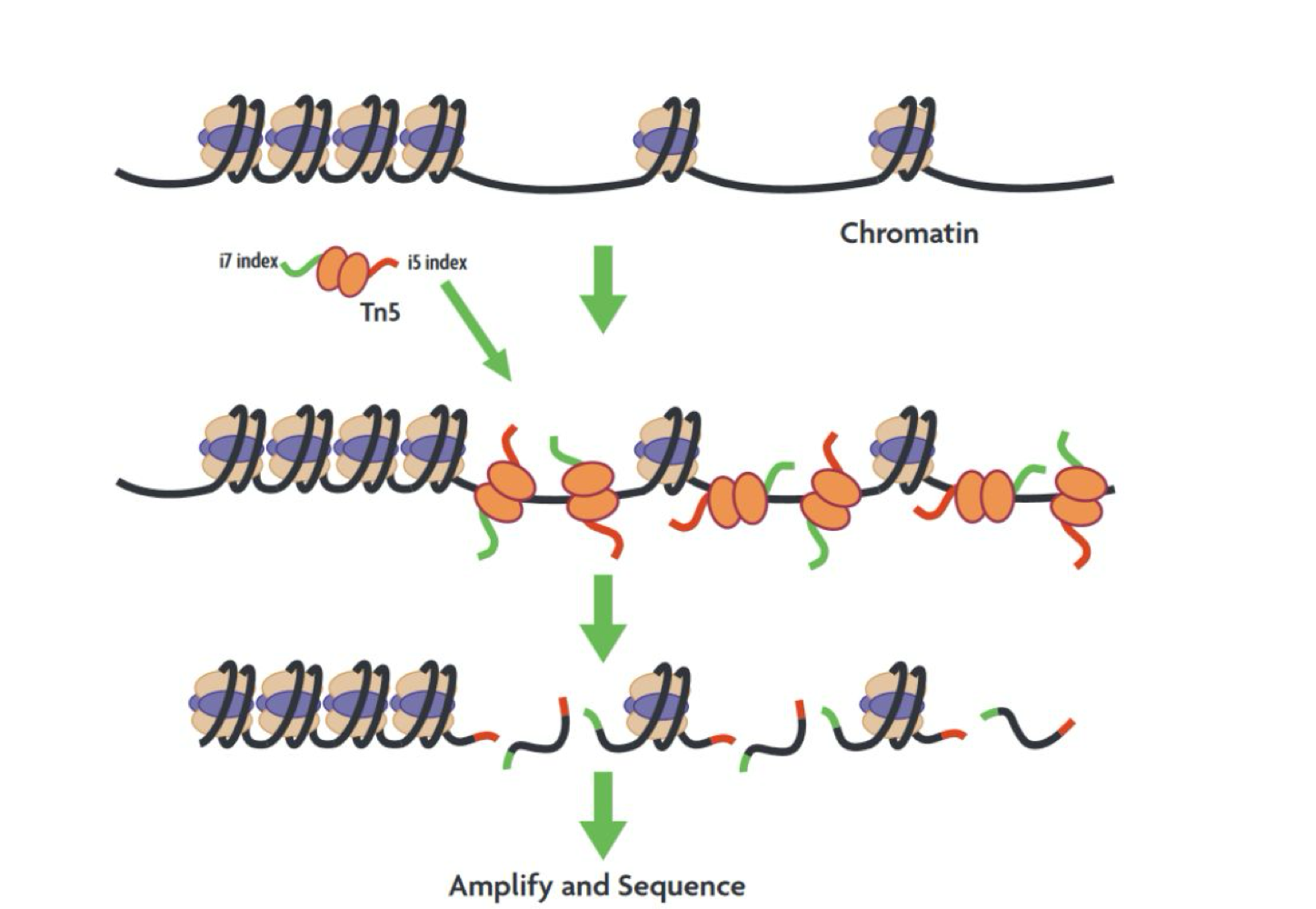
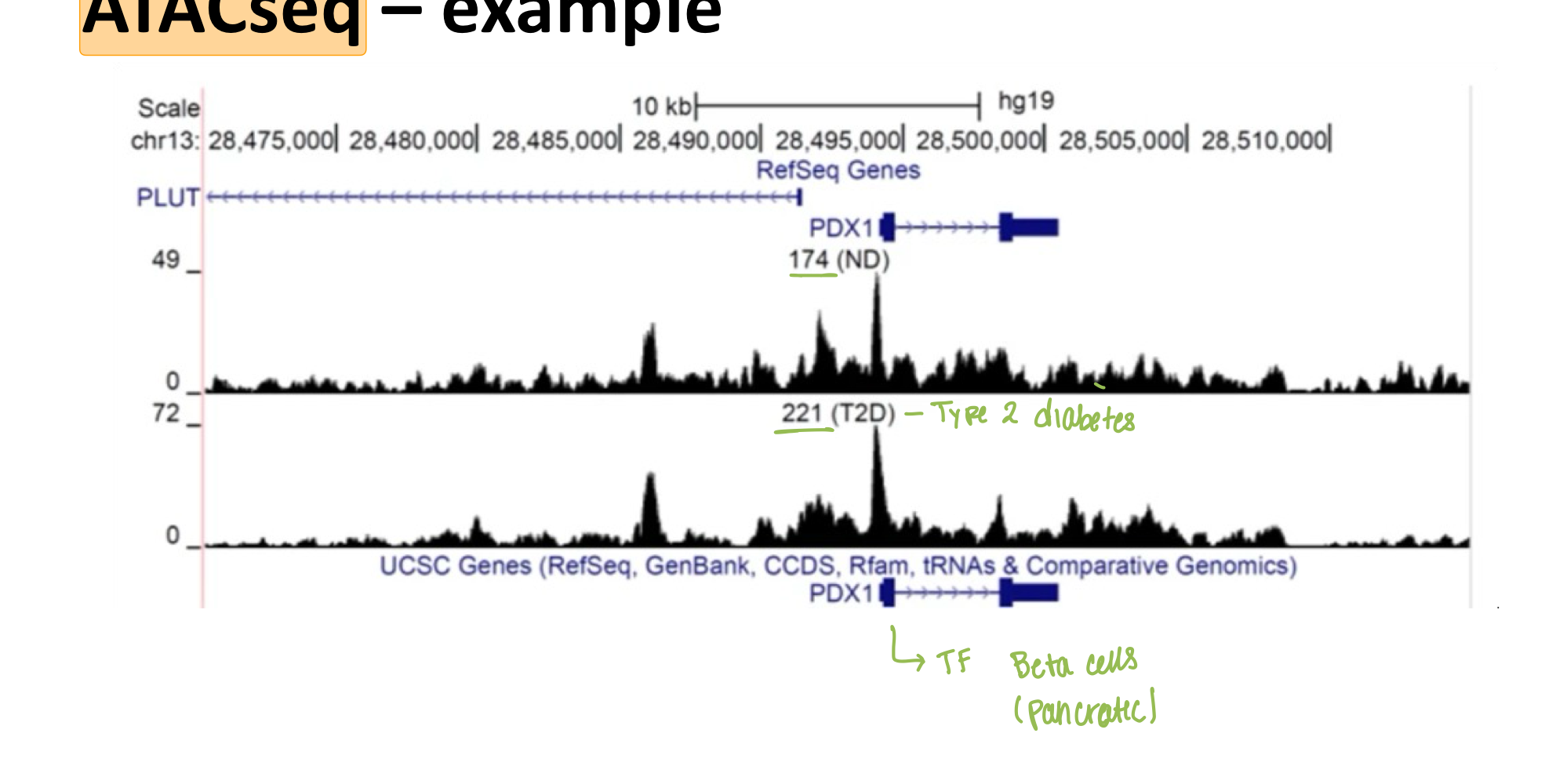
What does ATAC-Seq identify?
ATAC-Seq identifies open chromatin across the genome, with peaks at enhancers and promoters

What enzyme is used in ATAC-Seq?
Tn5 transposase cuts and inserts adapters into open chromatin
Where do you see peaks in ATAC-Seq?
Peaks correspond to locations of open chromatin (promoter regions)
What does EMSA detect?
EMSA (Electrophoretic Mobility Shift Assay) detects protein-DNA interactions, specifically binding of TFs to enhancers
What causes a "shift up" in EMSA?
A shift up occurs when protein binds to DNA
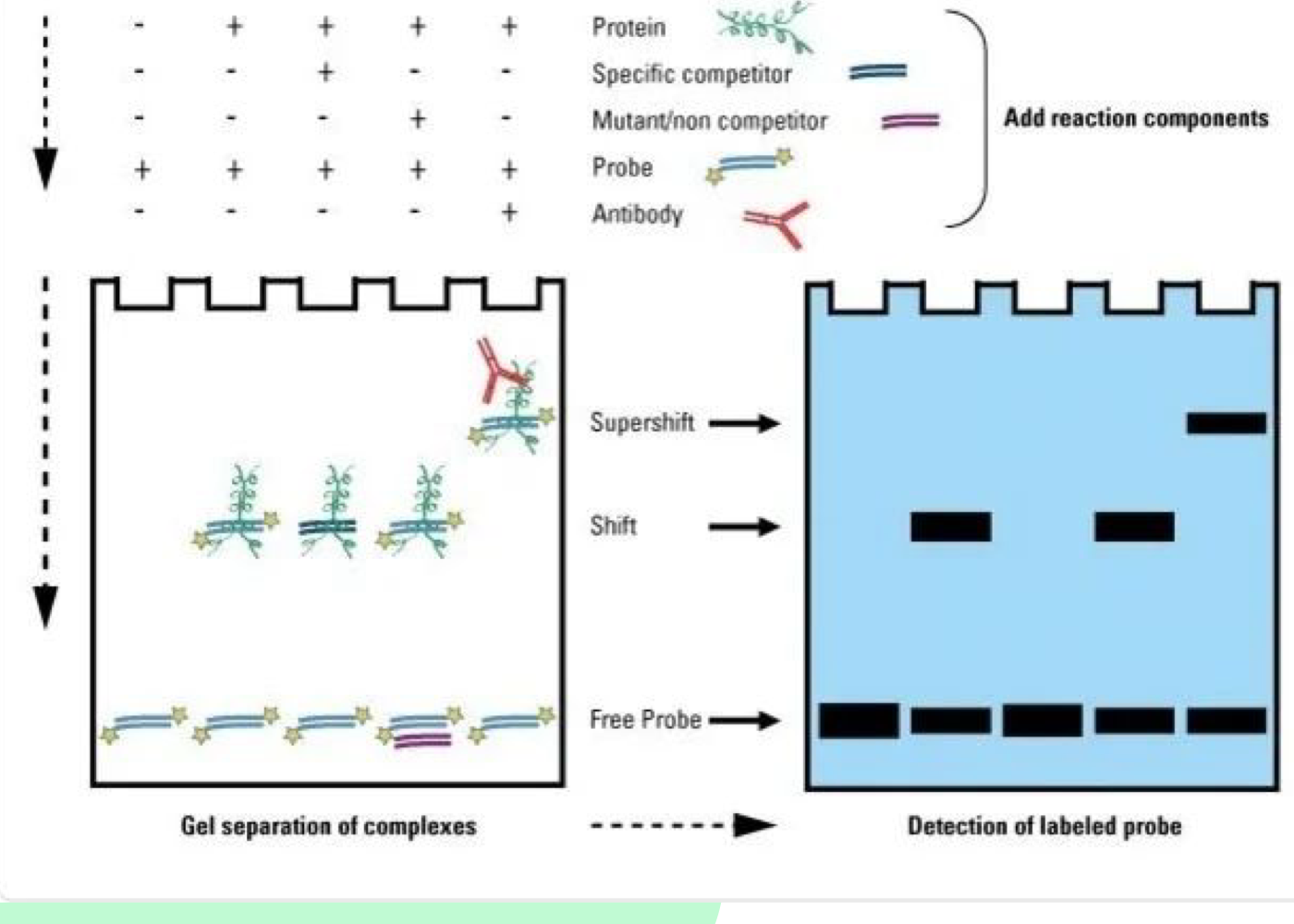
What is a supershift in EMSA?
A supershift occurs when an antibody specific to the protein-DNA complex is added, confirming that the protein is binding to the DNA of interest
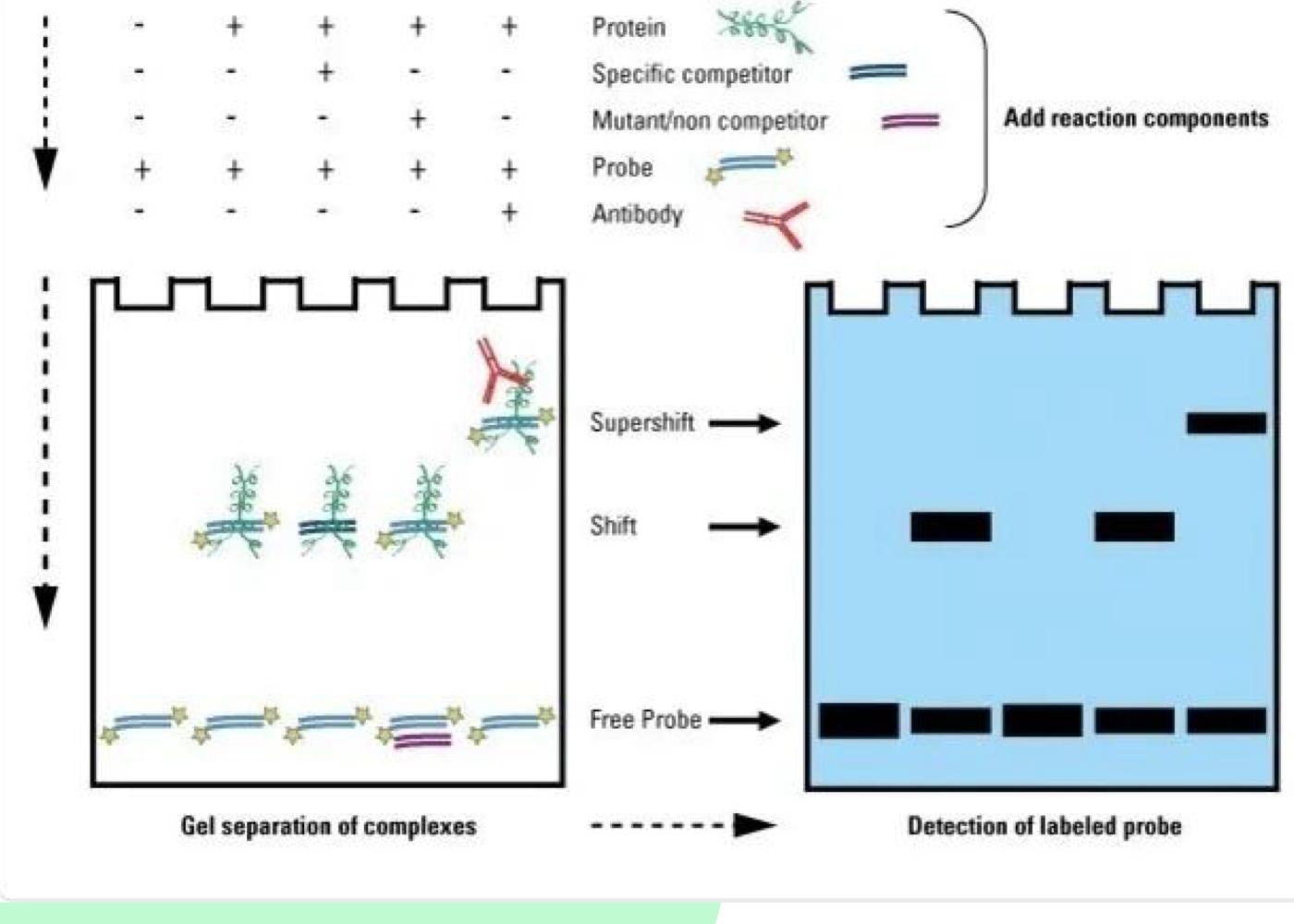
What are the components needed for EMSA?
Protein of interest, tagged DNA sequence, unlabeled DNA (competitor), and non-competitor sequence (DNA without specific binding site)
What do enhancer reporter assays identify?
They identify and characterize enhancer DNA sequences
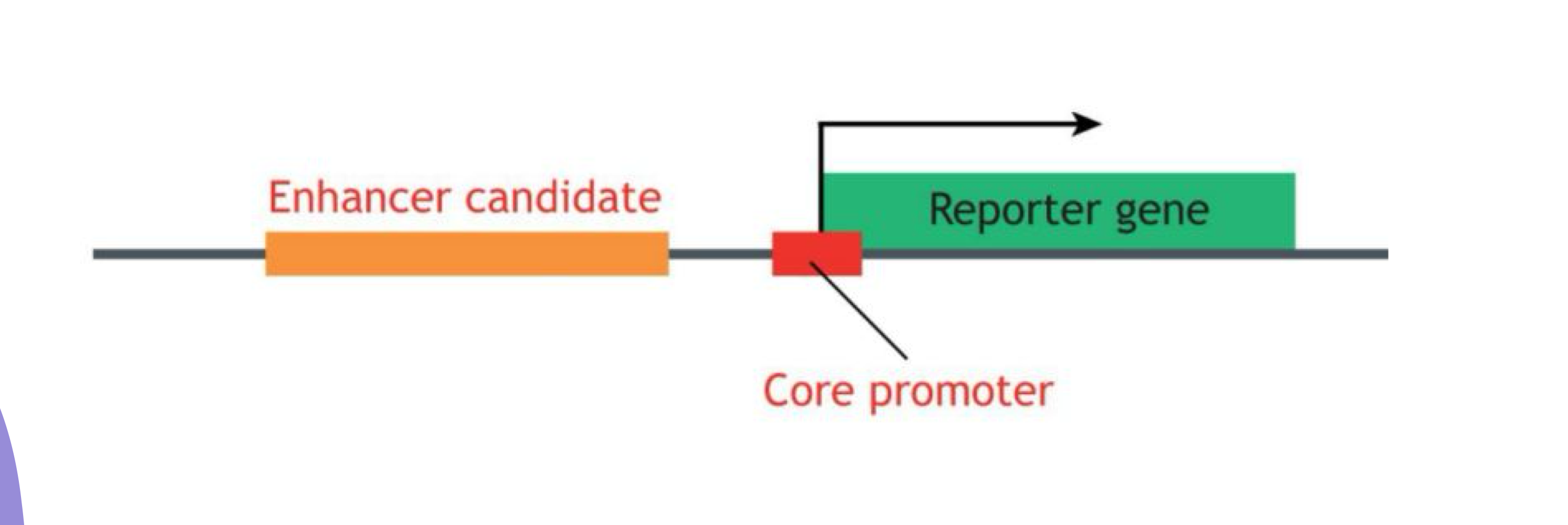
What are the components of an enhancer reporter construct?
Enhancer + Core promoter + Reporter gene (e.g., GFP)
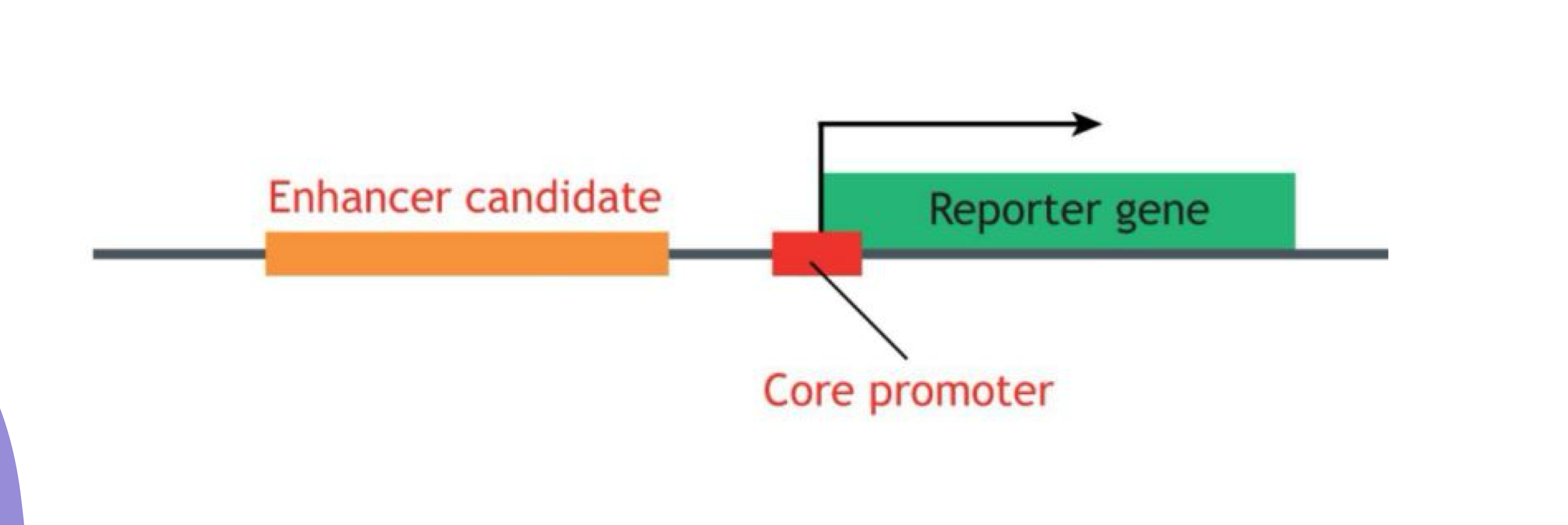
What indicates that an enhancer is sufficient in reporter assays?
Reporter gene expression indicates the enhancer is sufficient
How does DNA methylation affect gene expression?
DNA methylation inhibits transcription and gene expression
Methylates cytosines to 5-methyl-cytosine

How does DNA methylation prevent TF binding?
It attracts methyl CpG binding proteins that block TF binding and trigger heterochromatin formation
Where does DNA methylation typically occur?
Highly repetitive elements, non-coding DNA regions, and CpG islands (cytosine-phosphate-guanine regions near promoters)
What do DNMT3a/b do?
DNMT3a/b perform de novo methylation during embryogenesis on unmethylated DNA (cytosine)
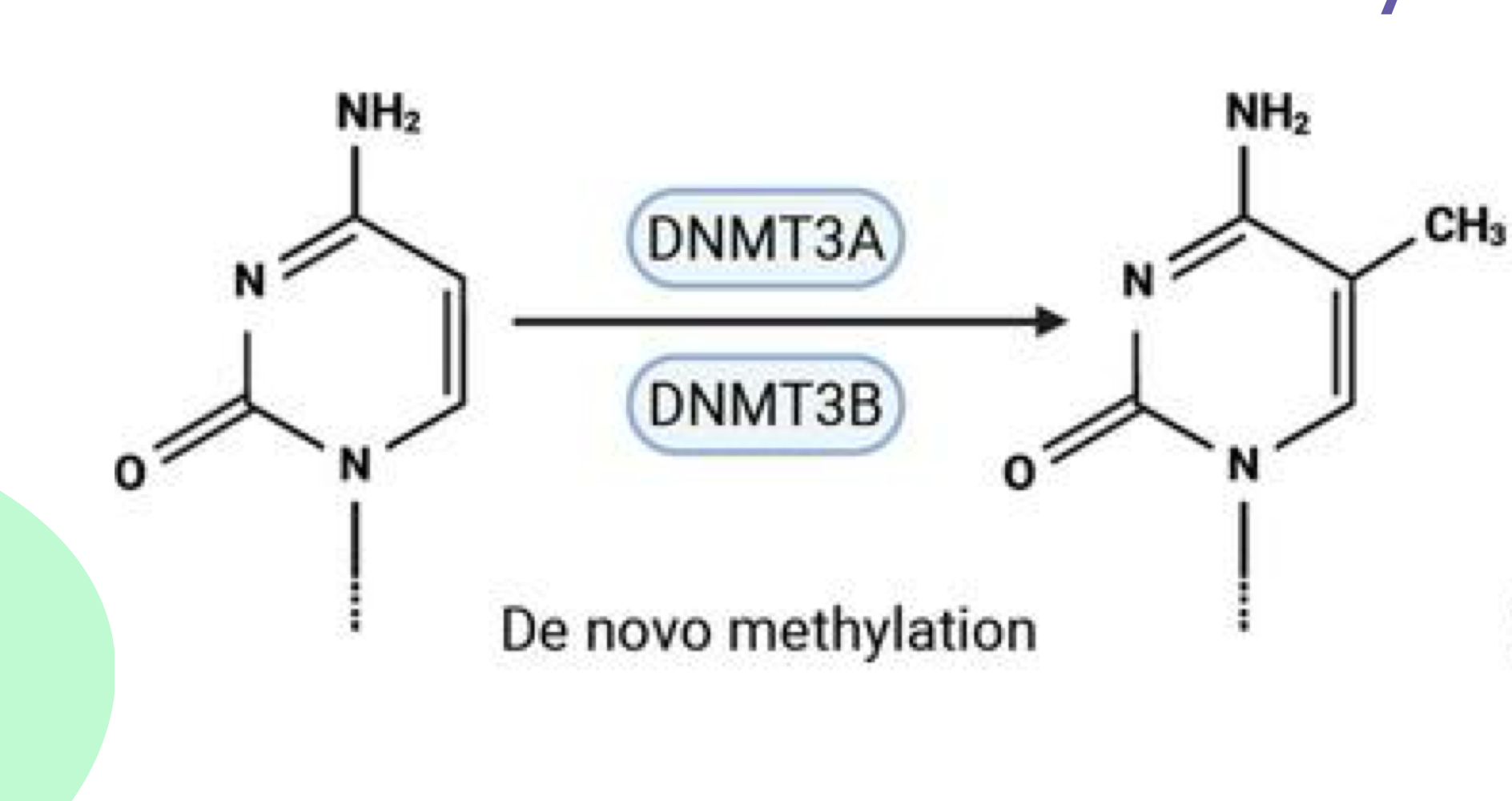
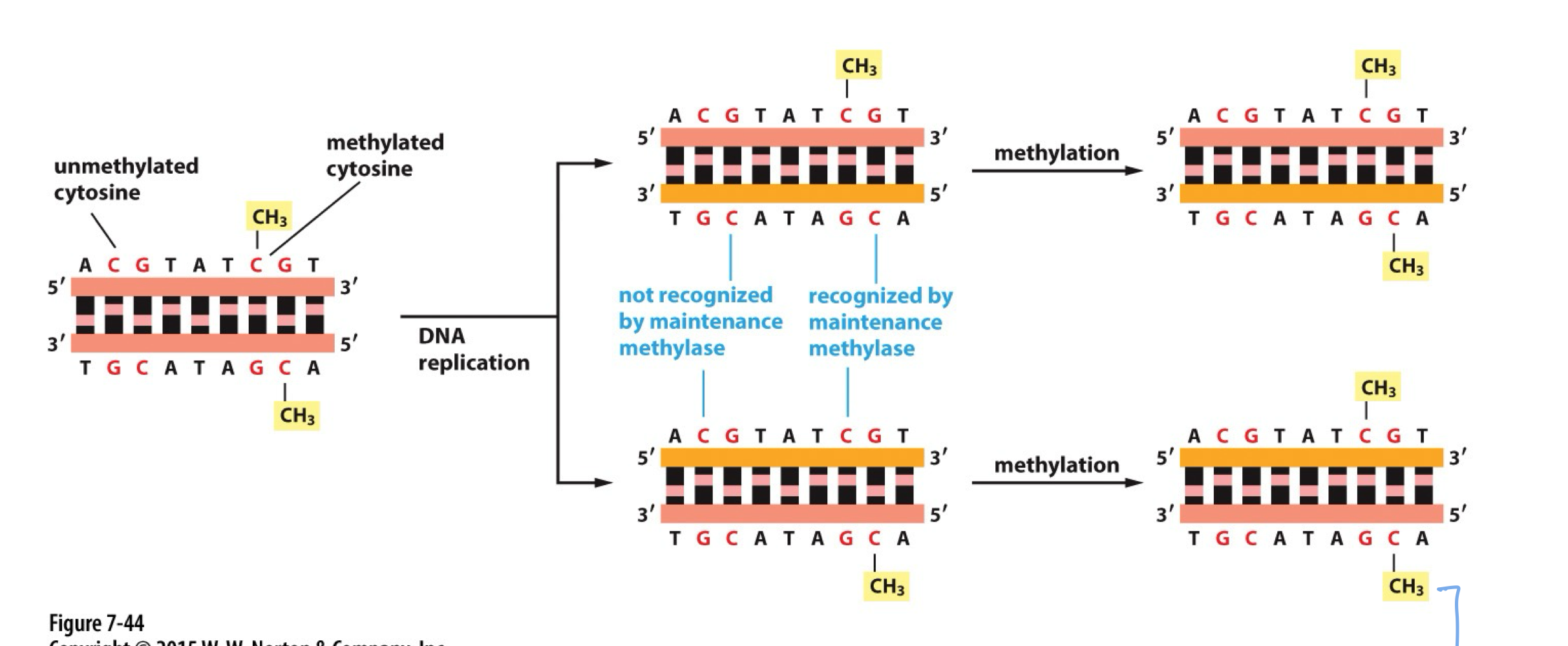
What does DNMT1 do?
DNMT1 performs maintenance methylation throughout life and replication on partially methylated DNA
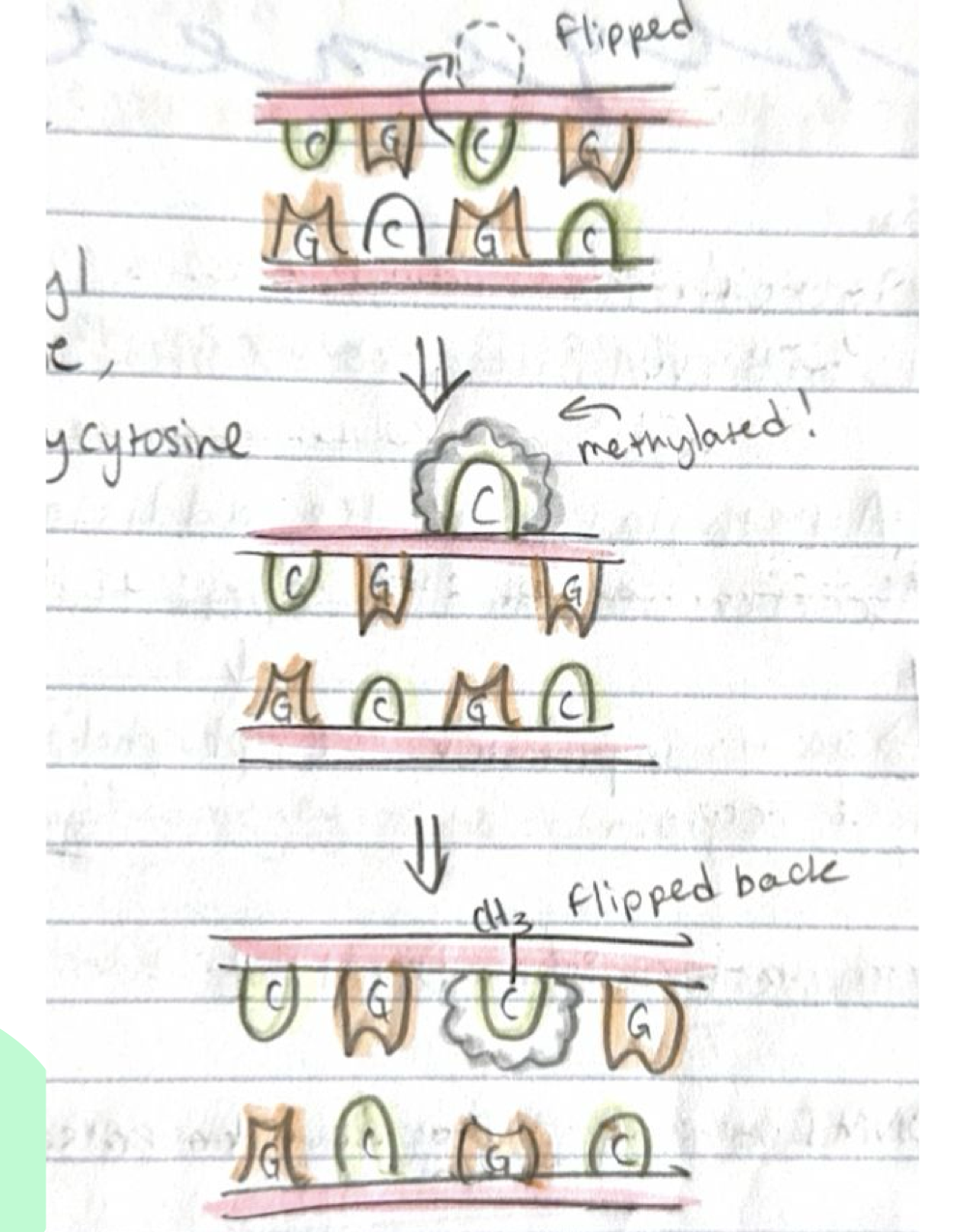
How do DNMTs add methyl groups?
DNMTs add a methyl group at the 5th position of cytosine, producing 5-methylcytosine. The process "flips" cytosine up to attach the methyl group then back down.
How does TET cause demethylation?
ET adds a hydroxy group (-OH) to 5-methylcytosine to make 5-methylhydroxycytosine.
This leads to demethylation through two pathways:
1) Replication-dependent dilution - the hydroxy group prevents DNMT1 from recognizing 5mC, so methylation is not maintained and is diluted after several rounds of replication
2) Thymine DNA glycosylase - a DNA repair mechanism that excises oxidized 5mC and repairs the DNA

How does impaired DNMT1 lead to demethylation?
If DNMT1 cannot methylate partially methylated DNA, the cytosine will lose its methylation in subsequent rounds of replication. This can also occur when TET adds hydroxy groups to 5-methylcytosine, preventing DNMT1 recognition during replication-dependent dilution
What is the effect of demethylation on gene expression?
Demethylation generally increases gene expression by removing repressive methylation marks
What is euchromatin?
Euchromatin is open, transcriptionally active chromatin
What histone marks are associated with active chromatin?
H3K4me3 and H3K9ac are associated with active chromatin
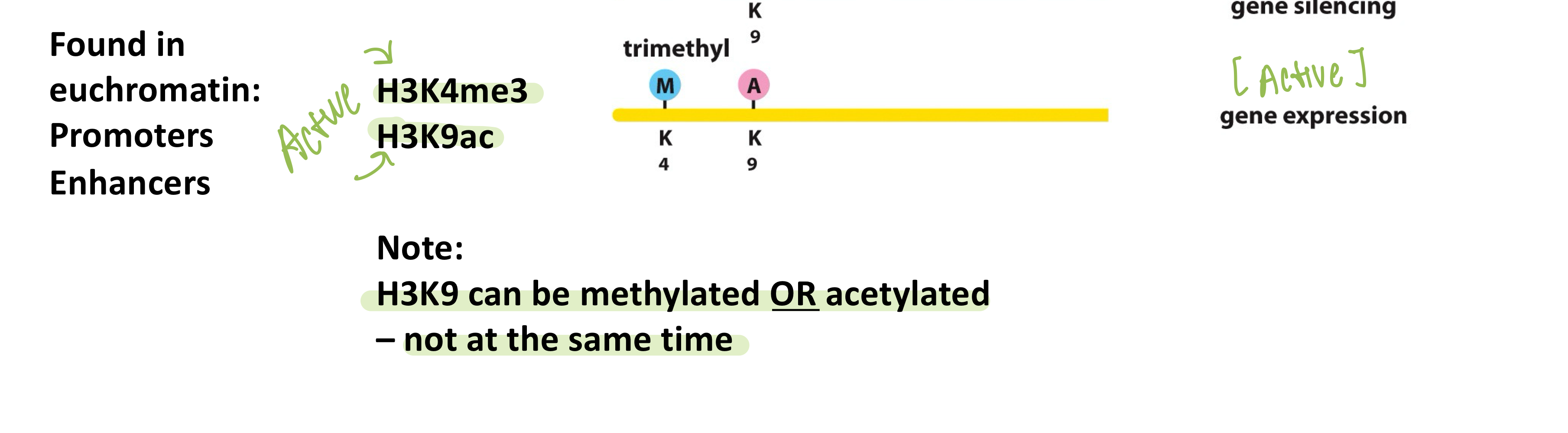
What is heterochromatin?
Heterochromatin is condensed, transcriptionally inactive chromatin
What is constitutive heterochromatin?
Constitutive heterochromatin is ALWAYS condensed and inactive (found at centromeres, telomeres, and repetitive regions)
What histone mark is associated with constitutive heterochromatin?
H3K9me3 is associated with constitutive heterochromatin (repressive)

What is facultative heterochromatin?
Facultative heterochromatin contains conditionally silenced genes that may be active at different developmental states

What histone mark is associated with facultative heterochromatin?
H3K27me3 is associated with facultative heterochromatin (repressive)

What characteristics are found in expressed gene regions?
Open chromatin, H3K4me3, H3K9ac, transcription factor binding, and low DNA methylation
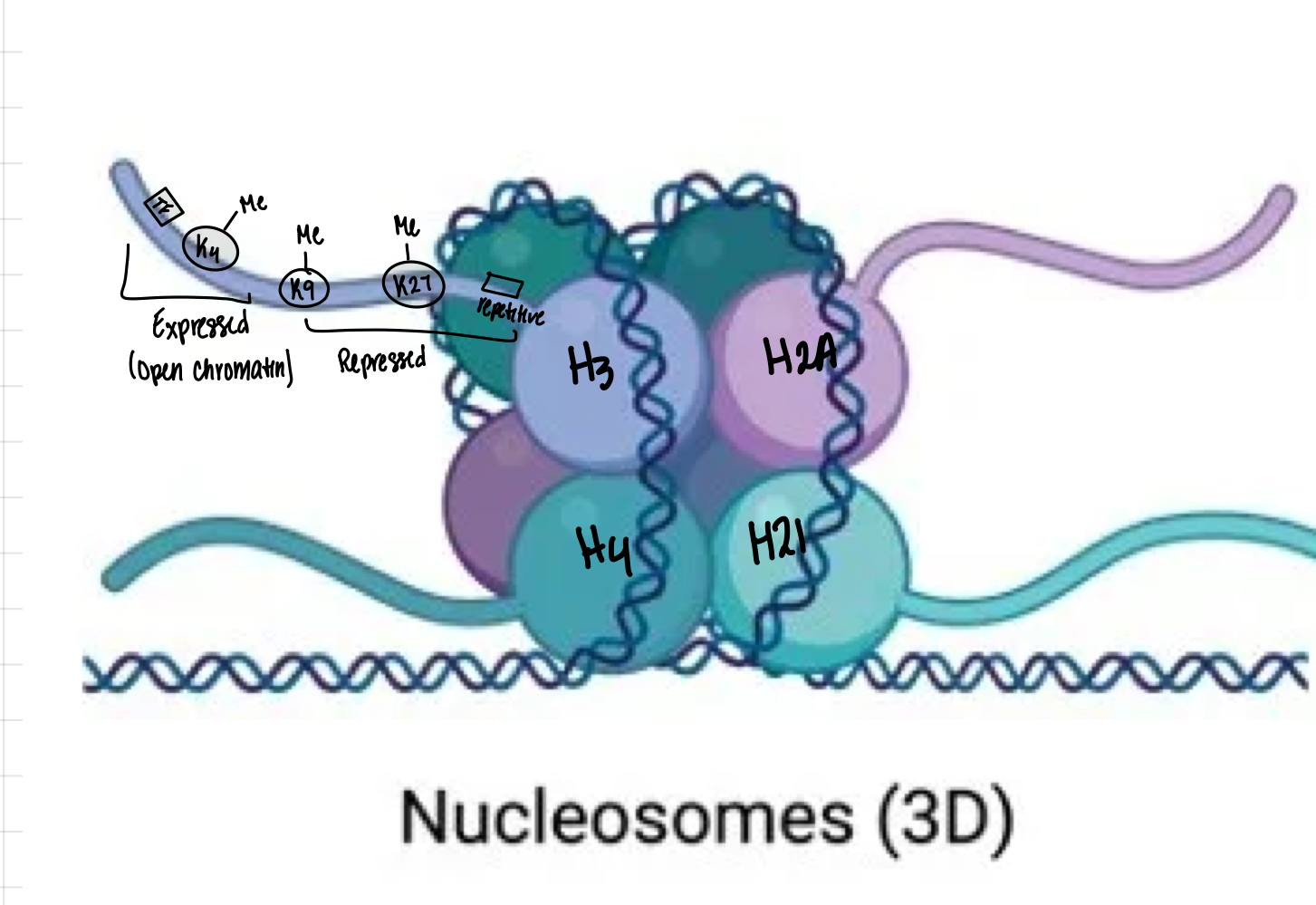
What characteristics are found in repressed gene regions?
Condensed chromatin, H3K27me3 or H3K9me3, and DNA methylation
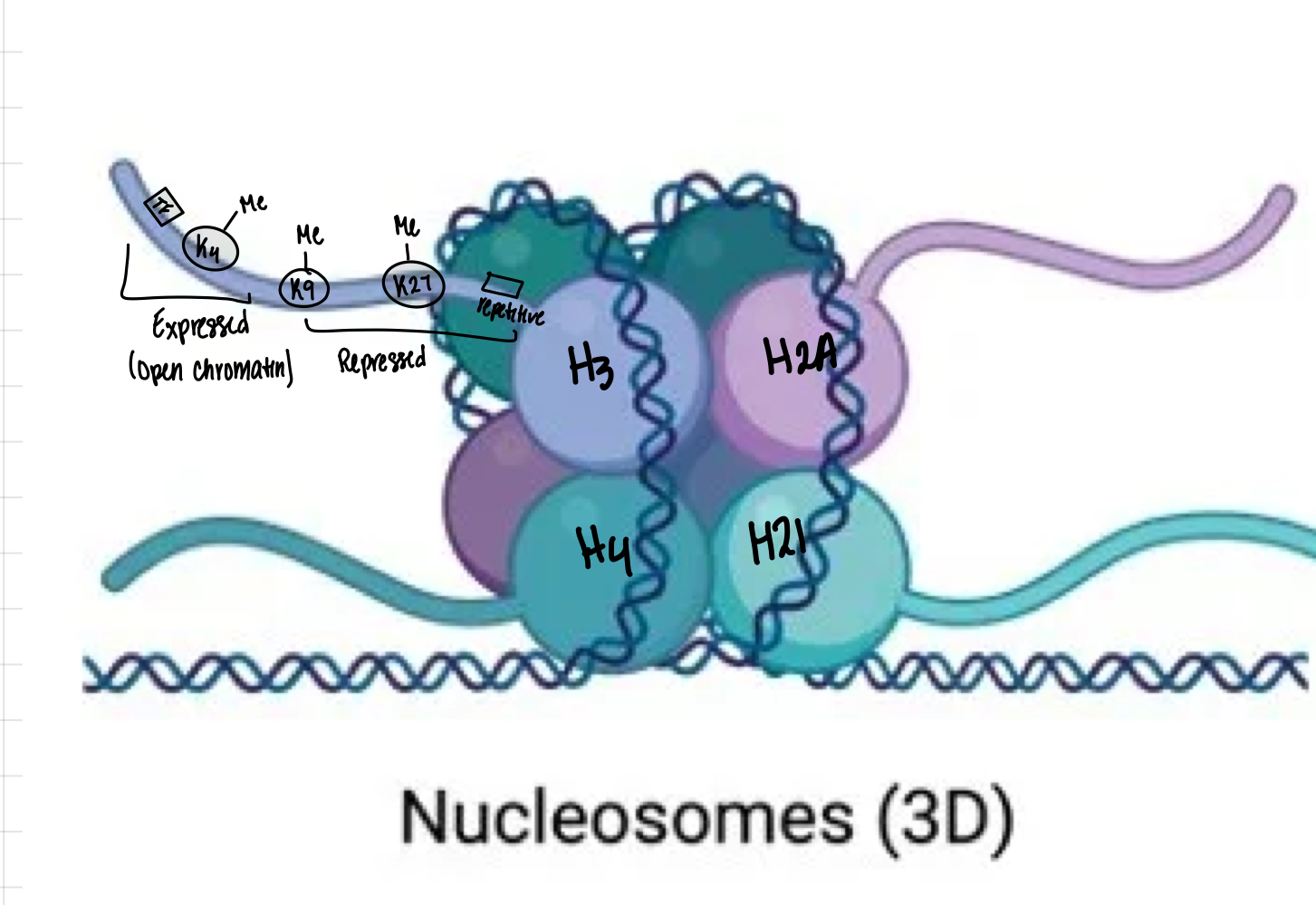
Where would you find H3K9me3?
H3K9me3 is found in constitutive heterochromatin regions like centromeres, telomeres, and repetitive regions
Which histone modifications spread to neighboring nucleosomes?
Repressive marks H3K27me3 and H3K9me3 spread to neighboring nucleosomes
Which histone modifications do NOT spread?
Active marks H3K4me3 and H3K9ac do NOT spread
Types of post-translational histone modification and their effects on gene expression
Methylation: Adds a methyl group (can be activating/inactivating)
Acetylation: Adds an acetyl group (activating)
Phosphorylation
Ubiquitination