Genetics ch 14
1/105
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
106 Terms
what is a peptide bond
is the covalent bond between amino acids in a polypeptide, that forms between the carboxyl end of an amino acid and the amino end of the next
what forms between the carboxyl end of an amino acid and the amino end of the next
peptide bond
where is calmodulin present in
all eukaryotic cells
what is a calmodulin
It is a small protein that consists of a single polypeptide, and has tertiary structure. The helical portions are alpha helices.
What is an alpha helices
a type of secondary structure observed in many proteins
what do many proteins consist of
a single polypeptide
ex. calmodium
what is an exampled of a protein with multiple polypeptides
Hemoglobin (Hb)
what is hemoglobin (Hb)
is the protein inside the red blood cells that carries oxygen in the bloodstream.
4 polypeptides
2 a-globins
2 b-globins
has quaternary structure
what is the primary structure of a protein
its sequence of amino acids
Secondary structure of a protein
interactions between amino acids cause the primary structure to fold into a secondary structure
tertiary structure of proteins
the secondary structure folds further into a tertiary structure
quaternary structure
two or more polypeptide chains may associate to create a quaternary structure
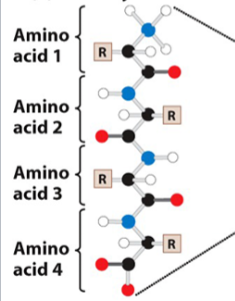
What is this
a primary structure of a protein
polypeptide: sequence of amino acids
1ry structure
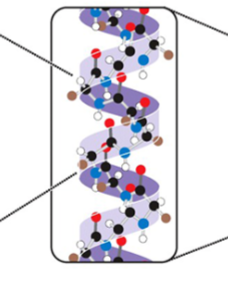
what is this
Secondary structure of a protein
Alpha helix: partial folding
2ry structure
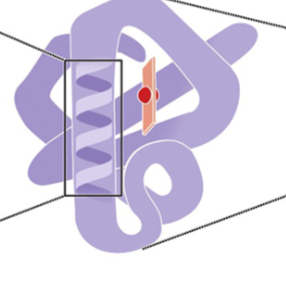
what is this
Tertiary structure of a protein
Globin: complete folded polypeptide
3ry structure
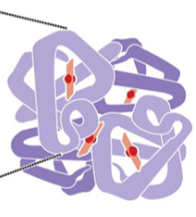
what is this
Quaternary structure of a protein
Hemoglobin
4ry structure
Sickle cell anemia
a single amino acid substitution in B-globin reduced the affinity of Hb for O2 and impairing the solubility and absorption of oxygen.
the polypeptide forms large polymers that distort the shape of red blood cells, affecting blood flow and oxygen transport
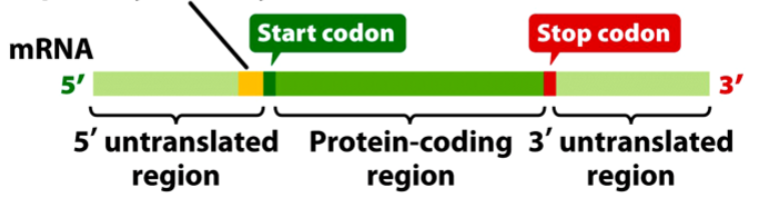
What is a codon
a three-base (nucleotides) sequence of DNA (a triplet) that codes for an amino acid
The genetic code is usually expressed on the table as what
mRNAs in the 5 → 3 orientation
Start codon
AUG → also codes for methionine
Stop codons
UAA, UAG, UGA (these do not code for any amino acids)
What is a reading frame
a linear sequence of codons in a nucleic acid defined by a start codon and ending with a stop codon
What does degenerate genetic code mean
multiple codons can code for the same amino acid
codon for methionine (Met)
AUG (start codon as well)
what are the characteristics of the genetic code
unambiguous
degenerate
universal
commaless
non-overlapping
Unambiguous
each of the 61 triplets code for only one of the 20 amino acids
degenerate
most amino acids are encoded by more than one codon
Universal
most living organisms use the same code, but exceptions exist
Commaless
there are no breaks between the codons in the reading frame (all the bases of the translated sequences are part of codons)
non-overlapping
in triplets in a reading frame are in a tandem sequence and do not overlap
what is the flow of genetic information
DNA → mRNA → protein
(transcription → translation)
How do the template strand and mRNA relate
The template (antisense) DNA strand:
runs 3 → 5
Used by RNA polymerase to make mRNA
mRNA is complementary to this strand
example: DNA 3 - TACCACAACTCG - 5
mRNA strand
runs 5 → 3
sequence matches the DNA coding (sense) strand except U replaced T
example: mRNA 5 - AUGGUGUUGAGC - 3
Translation
ribosome reads mRNA codons (triplets)
AUG = start codon → met
each codon → one amino acid
example protein
met - val - leu - ser
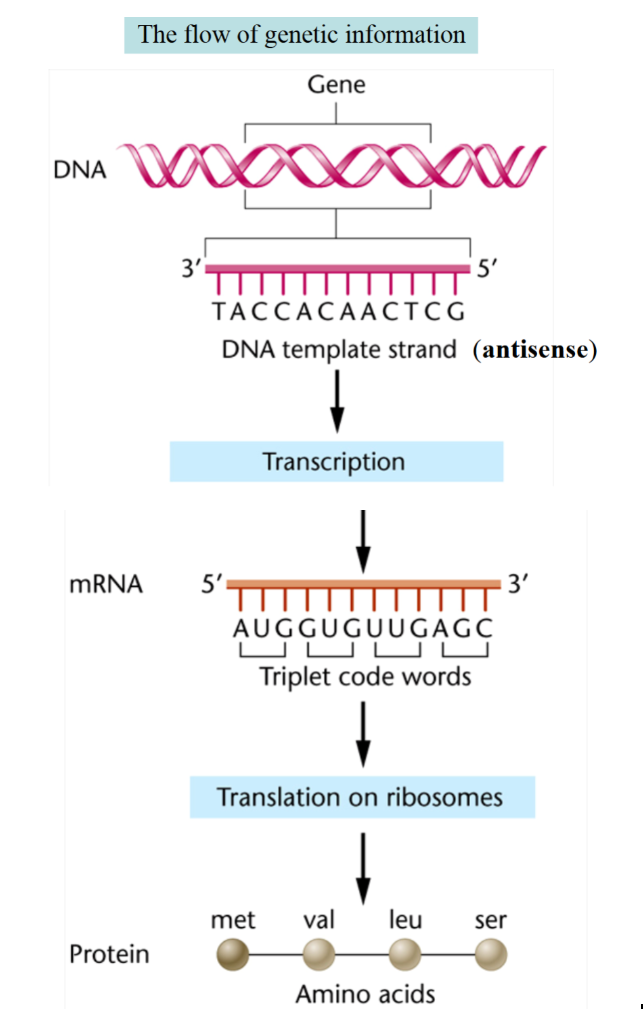
what is this
the flow of genetic information
what can spontaneous mutations result in
permanent changes
What causes spontaneous mutations
they are not caused by errors in DNA replication. DNA exists inside of the cell, which is an open system. DNA is then exposed to radiations and molecules that can react with it and cause chemical alterations.
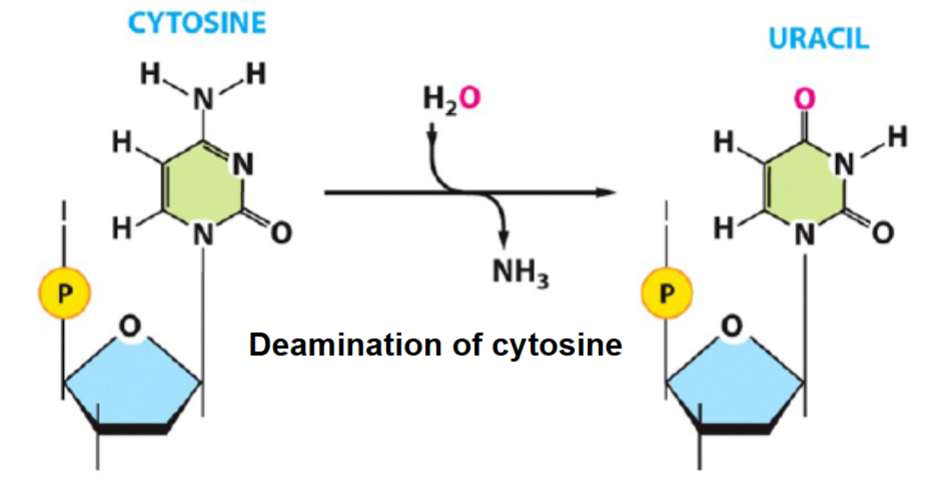
What is this
the deamination of cytosine
spontaneous mutations
original C-G base pair and then after deamination → U-A
The result in a C → T transition mutations after replication
to fix: the cells use uracil-DNA glycosylase (base excision repair) to remove uracil from DNA
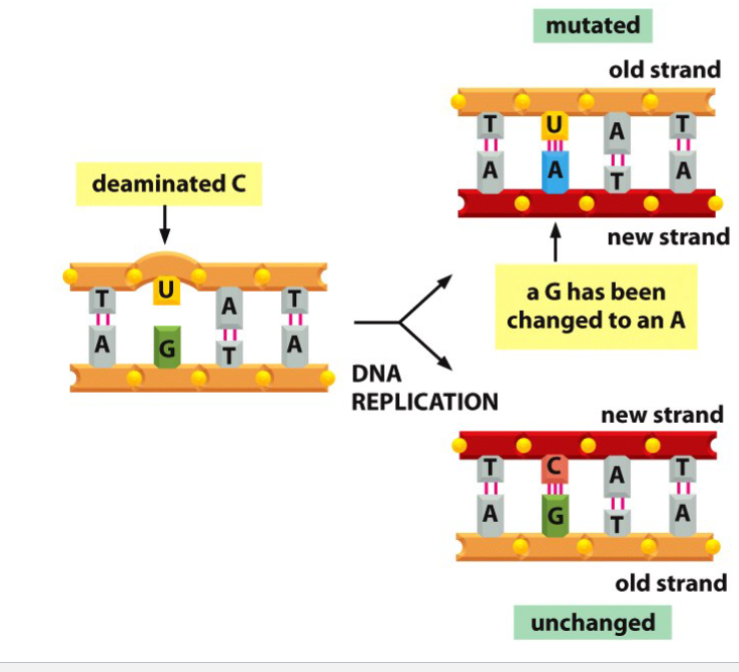
What is this
a spontaneous mutation resulting in a permanent base pair substitution in the top right as G has been turned to A to pair with U, and the bottom right has transformed C back to U and G stays the same.
When a base is replaced by a different base in DNA, the result is a permanent single codon change
substitution mutations in protein coding genes
Type of substitution mutations in protein-coding genes
silent
missense
nonsense
what is a silent substitution mutation
the resulting new codon codes for the same amino acid as the original codon
what is a missense substitution mutation
the new codon codes for a different amino acid
what is a nonsense substitution mutation
the new codon is a stop codon, resulting in a shorter polypeptide
what might cause mutations
errors in DNA replication
where do mutations occur and where may they be copied to
they occur in DNA and may be copied to mRNAs
example of a silent mutation
wild type codon: AGU → ser
mutation: AGC (U→C)
amino acid stays Ser
example of a missense mutation
wild type: AGU → ser
Mutation: AAU → Asn (G→ A)
This changes the amino acid (ser → asn)
Example of a nonsense mutation
wild type: AGU → ser
mutation : UGA → stop (A→ U)
translation ends early → truncated protein
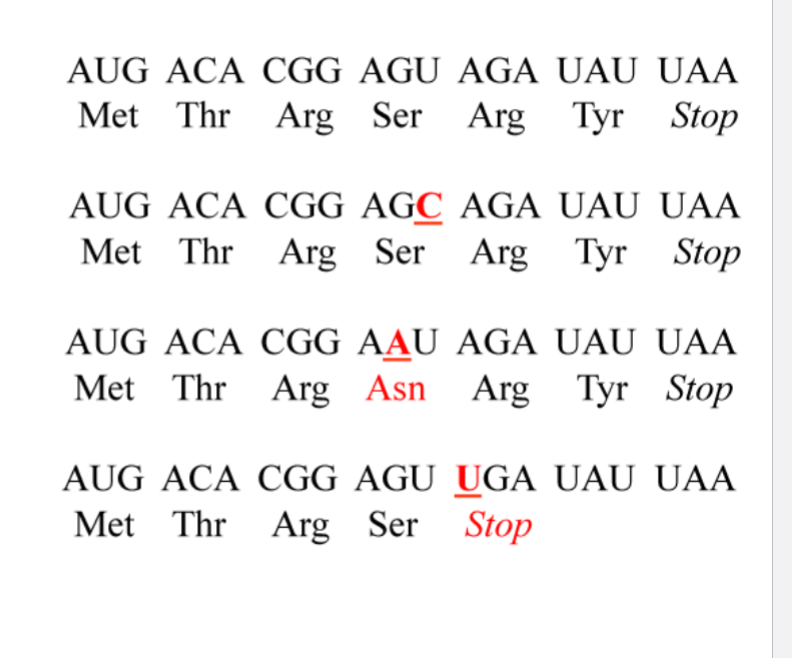
What is the wildtype
mRNA: AUG ACA CGG AGU AGA UAU UAA
Protein: Met - Thr - Arg - Ser - Arg - Tyr - Stop
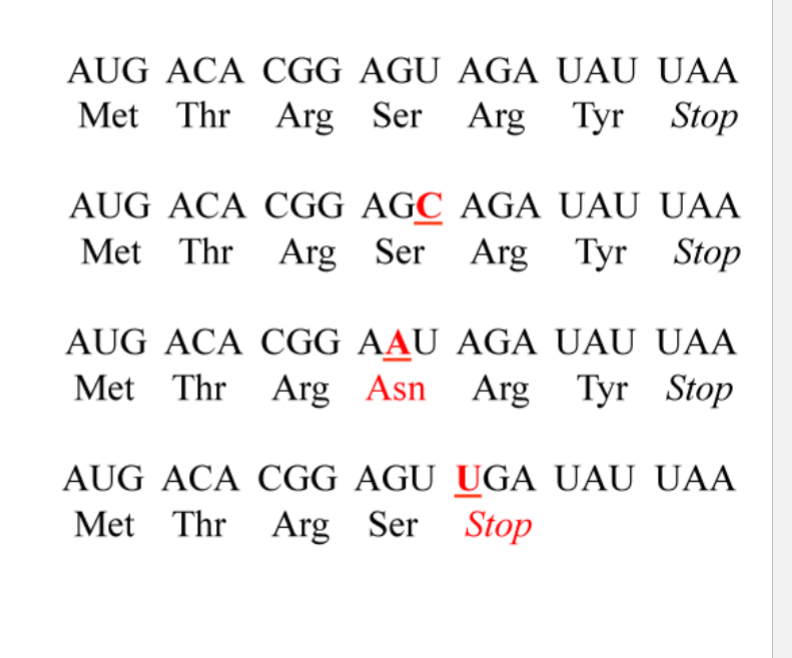
what is the silent mutation
AGG (ser) was AGU (ser)
U → C

What is the missense mutation
AAU (asn) was AGU (ser)
G → A
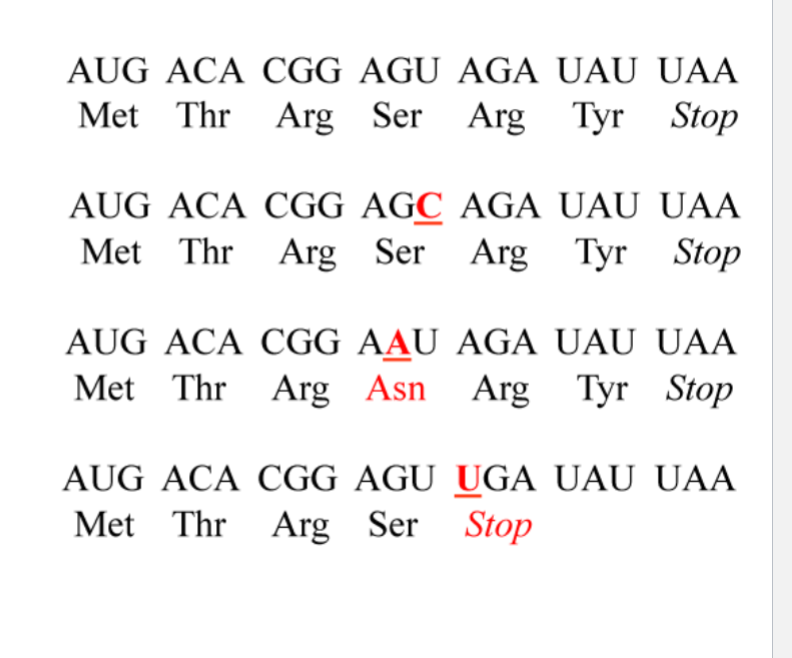
What is the nonsense mutation
UGA (stop) was AGU (ser)
A → U
what does the insertion of a single base result in
a frameshift mutation
what are insertion due to
errors in DNA replication
all the codons from the insertion site on are changed, resulting in frameshift
the deletion of single bases by errors in DNA replication also occur and also result in frameshift mutations
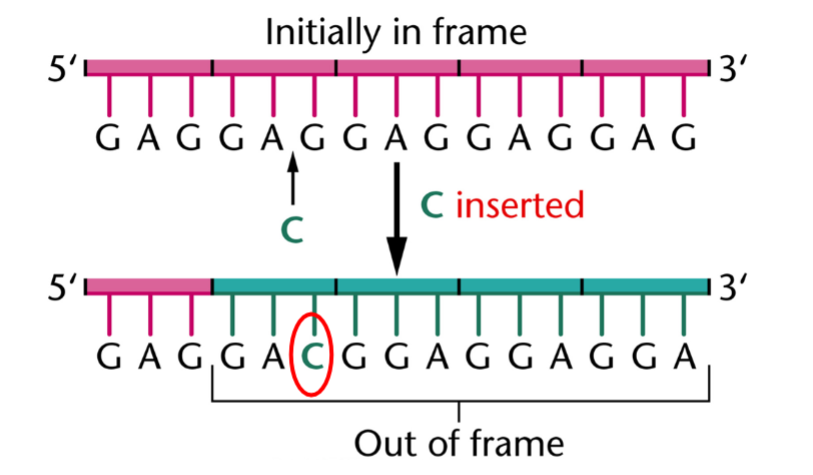
what is this
the insertion of a single base resulting in a frameshift mutation
What is huntington’s disease
A neurodegenerative disorder
an autosomal dominant disease caused by the expansion of trinucleotide CAG repeats, which code for glutamine in the mRNA, in the huntingtin gene, resulting in polyglutamine huntingtin proteins
what does the number of CAG repeats mean
In trinucleotide repeat expansions the number of CAG repeats determines if someone has the phenotype disease
Number of CAG repeats phenotype
11-35 normal
36-39 borderline
40 and over disease
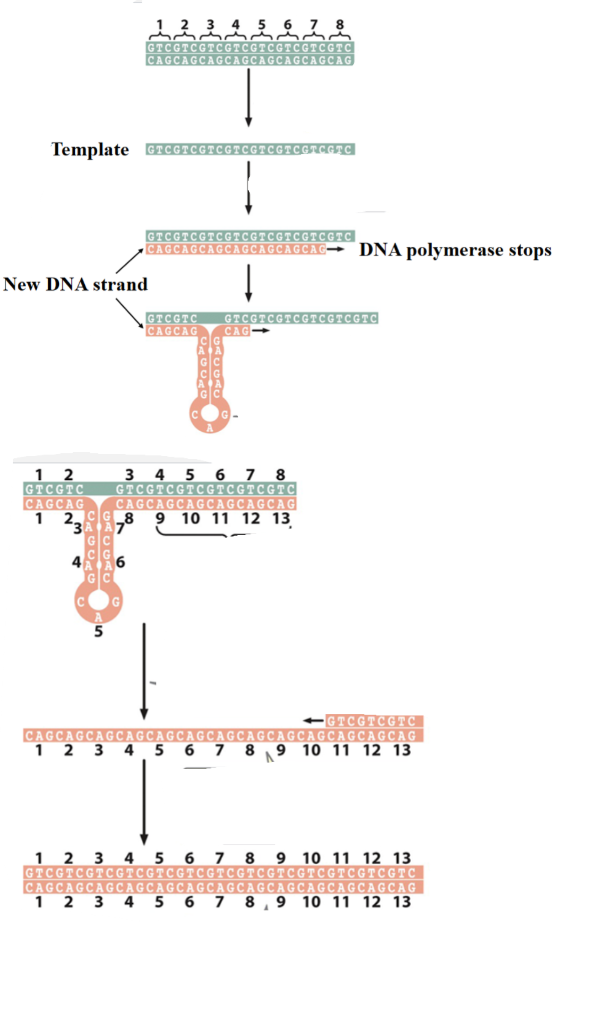
What is this explain
Trinucleotide repeats expansion
1) the DNA molecule has 8 copies of a CAG repeat
2) the two strands separate
3) and replicate
4) In the course of replication, a hairpin forms on the newly synthesized strand
5) causing part of the template strand to be replicated twice and increasing the number of repeats on the newly synthesized strand
6) the strand of the new DNA molecule separate
7) and the strand with extra CAG copies severs as a template for replication
8) The resulting DNA molecule contains 5 additional copies of the CAG repeat
Translation basic requirements
mRNA
charged transfer RNAs (tRNAs)
ribosome
other requirements: initial factors, elongation factors, and energy sources
no primers of any kind
What is this
Shine Dalgarno sequence in prokaryotes only

what is this
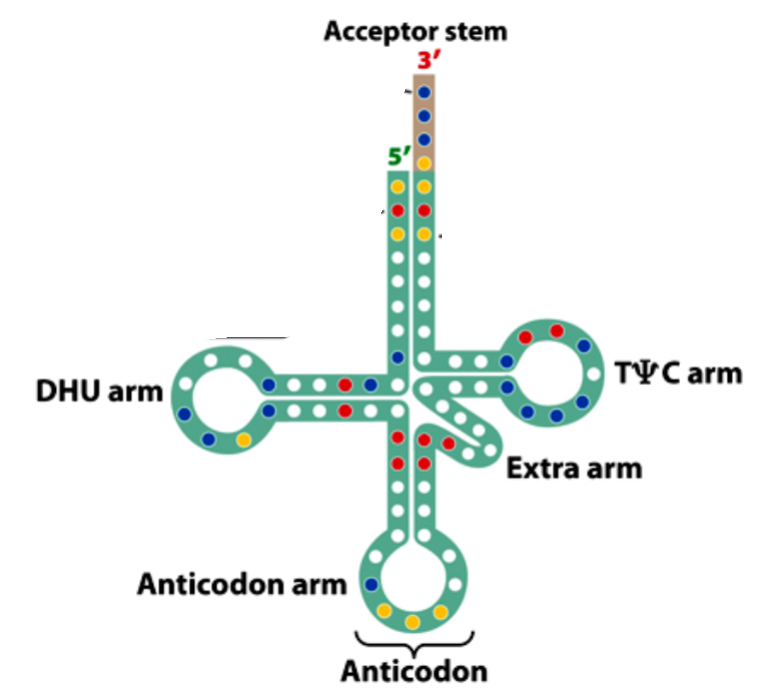
Transfer RNA (tRNA)
an amino acid is attached to the 3 end of the tRNA and is ALWAYS CCA
The anticodon loop base-pairs with a codon in an mRNA
The post-transcriptional modification of bases in tRNA result in what
unusual bases
what is inosine
is a modified adenine
what are unusual bases found in tRNAs
Inosinic acid (I)
1-Methyl inosinic acid (I^m)
1-Methyl guanylic acid (G^m)
NM-dimethyl guanylic acid (G^m)
Pseudouridylic acid
Ribothymidylic acid
what is the wobble hypothesis
The interaction between the third position of the codon in the MRNA and the first position of the anticodon in the tRNA is less critical and less constrained
some tRNA bases can pair with multiple mRNA bases at this position
this allows translation to occur without the need for the cell to synthesize all 62 tRNAs
inosine is a post-transcriptionally modified adenine that can occur at the wobble site of tRNAs
how many tRNAs are there
61
how many amino acids are there
20

what is this
degeneracy and the wobble hypothesis
During anticodon and codon base paring why are the codon and anticodon antiparallel
because tRNA binds mRNA in opposite orientations:
mRNA is read 5 → 3
tRNA anticodon binds 3 → 5
This ensures correct base pairing during translation
what is the wobble position
The 3rd base of the mRNA codon and 1st base of the tRNA anticodon
allowing flexible pairing, meaning one tRNA can recognize multiple codons
how do the base-pairing of the codon and anticodon pair
anti-parallel
what are the essential amino acids for humans
valine, leucine, isoleucine, phenylalanine, tryptophan, lysine, histidine, methionine, threonine
what is charging catalyzed by
20 different aminoacyl tRNA synthases
the enzymes recognize the amino acids by their R groups, and the tRNAs by their shapes and base sequences
energy is spent in this process in order to eventually make a new peptide bond in the ribosome
what is charging a tRNA
the carboxyl group of the amino acid is covalently attached to the 3 end of a tRNA

what is this
the charging of a tRNA
What is this
recognition of tRNAs by Aminoacyl tRNA synthases
1) positions in blue are the same in all tRNAs and CANNOT be used to differentiate among tRNAs
2) Positions red are important in the recognition of tRNAs by one synthetase
3) Positions in yellow are used by more than one synthetase
step 1 of charging of a tRNA
Amino acid activation:
the amino acid (AA) is converted to an aminoacyl adenylic acid (AA-AMP)
This is the energy-consuming step on the process of eventually making a new peptide bond
requires ATP

Step 2 of the charging of a tRNA
Charging:
the aminoacyl adenylic acid loses the AMP and the carboxyl group of the amino acid is attached to the 3 end of a tRNA
the result is an aminoacyl tRNA (a charged tRNA)

Ribosome
the large particle of rRNA and proteins where translation occurs
two subunits: small and large
what is the svedberg unit (S)
is not a measure of molecular weight, but a measure of the rate at which particles sediment in a centrifugal field
this rate depends on weight, shape, and size
this unit can be used for measuring large molecules or large cell components such as ribosomes and organelles
whare are the Svedberg values for prokaryotic ribosomes
small subunit: 30S
large subunit: 50S
complete ribosome: 70S
what sites in the ribosome are translation components
peptidyl (P)
aminoacyl (A)
exit (E)
What are the steps in the initiation of translation
1) mRNA binds to the small ribosomal subunit with the AUG codon positioned on the P site
2) f-Met-tRNA (in prokaryotes) binds to the AUG codon
3) the large ribosomal subunit joins the complex
this whole process requires GTP for a source of energy plus a series of initiation factors (IF proteins)

What is this
Prokaryotic translation initiation
during prokaryotic translation initiation which molecule binds to the small ribosomal subunit to prevent premature large-subunit binding
initiation factor IF-3
during prokaryotic translation initiation what amino acid does the first bacterial tRNA carry
formyl-methionine (fMet)
During prokaryotic translation initiation which factor brings the first tRNA (fMet-tRNA) to the P-site
IF-2 + GTP
During prokaryotic translation where does the first tRNA (fMet-tRNA) bind in the ribosome
The P-site
What happens when GTP is hydrolyzed during initiation
initiation factors dissociate, allowing the large subunit to join
What is the final product of initiation
a 70S initiation complex ready for elongation
Do eukaryotes use formyl-methionine
No → only bacteria + mitochondria/chloroplast use fMet
but eukaryotic organelles (mitochondria and chloroplasts) do
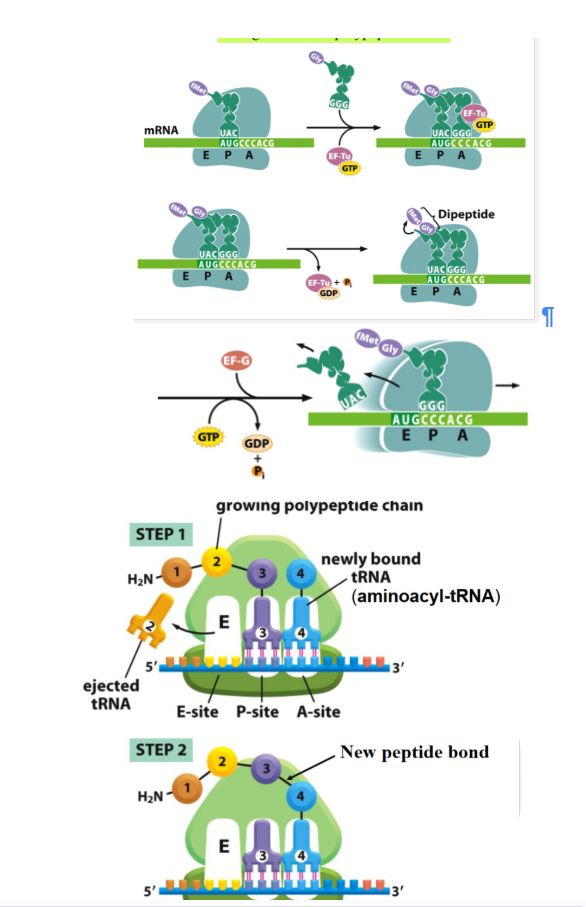
what does EF-Tu and GTP do during elongation
facilitate the binding of the second tRNA to the second codon at the A site
what forms during elongation when the amino acid on the first tRNA is transferred to the second tRNA
A peptide bond forms and dipeptide forms
during elongation Where does the first tRNA move after the peptide bond forms
to the E-site
during elongation what happens to the mRNA after the first peptide bond is formed
the mRNA is shifted to place the second codon in the P site and bring the third codon into the A site
during elongation what binds to the third codon
the tRNA carrying the third amino acid binds to the third codon on the A site
What forms during elongation when the dipeptide is transferred from the second tRNA to the third tRNA
a tripeptide attached to the third tRNA
What happens to the second tRNA after the tripeptide is formed
the second tRNA moves to the E site with the help of EF-G and GTP
Where does the third codon move after EF-G shifts the ribosome during elongation
the third codon moves to the p Site
after elongation steps what happens
translation continues
What is this
elongation of the polypeptide chain