m7 tot
1/65
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
66 Terms
Microtubule Structure
Comprised of 13 protofilaments
Arrayed circularly to form a tube wall
They’re staggered to resemble a spiral
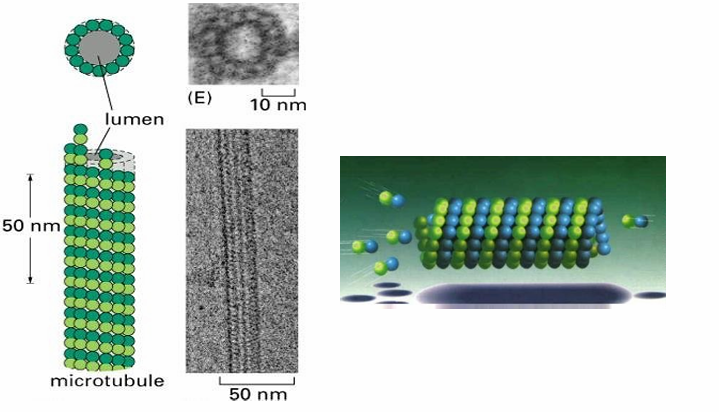
What are the basic subunits of each protofilament (microtubule structure)
Dimers of alpha and beta tubulin proteins
What are the GTP-binding properties of α- and β-tubulin subunits?
Both α- and β-tubulin bind GTP.
α-tubulin: GTP is tightly bound
never hydrolyzed
does not exchange with free nucleotides.
β-tubulin: GTP is loosely bound
hydrolyzed to GDP
exchanged for GTP in the cytosol.
How are tubulin subunits added and removed during microtubule assembly?
α- and β-tubulin subunits are added/removed as dimers.
αβ–GTP dimers have a higher affinity for the growing microtubule (more stable).
αβ–GDP dimers have a lower affinity and tend to dissociate from the filament.
Microtubule Polarity
They’re polar so the two ends have different characteristics and dynamics
(+) end = fast growing
(-) = slow growing
Within the dimers
the beta-subunit is closer to (+)
the alpha-subunit is closer to (-)
Microtubule Dynamics
Dimers with αβ–GTP are added to (+) end
Rescue phase
Dimers with αβ–GDP are released from shrinking filament
Catastrophe
GTP hydrolysis occurs within polymerized microtubule
Most of it consists of dimers containing αβ–GDP
(+) has GTP cap (unhydrolyzed) which favours growth
αβ–GTP dimers have a 4x slower disassociation rate in comparison to αβ–GdP
They thus have higher affinity for their neighbours and stay together
(+) end has dynamic instability
Oscillates between growth or shortening
High [GTP-tubulin] = polymerization
Low [GTP-tubulin] = depolymerization
EB1 Protein (Microtubule)
Plus-end binding protein
Prevents premature catastrophes
Acts as positive regulator of microtubule growth
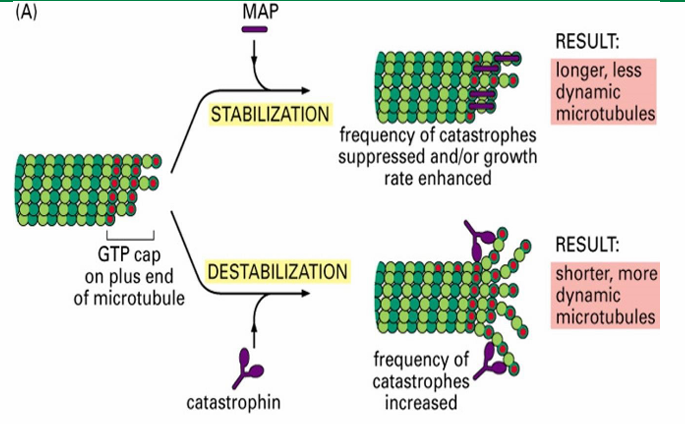
MAPs - Microtubule Associated Proteins
Proteins controlling the assembly and disassembly of microtubules
MAPs - Microtubule Associated Proteins (Function)
Interconnect microtubules to form bundles
Inc stability
alter rigidity
influence assembly rate
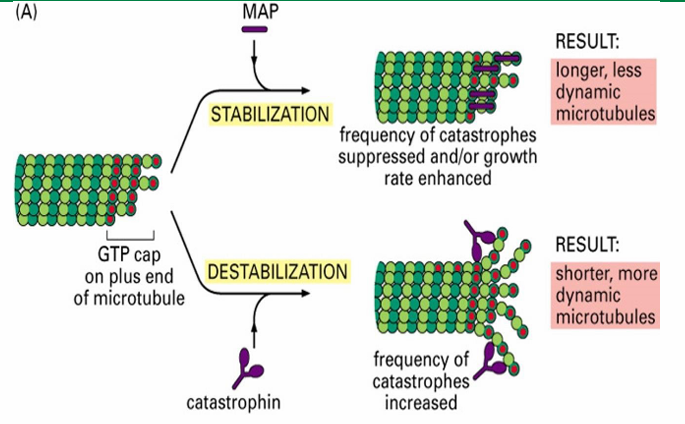
MAPs - Microtubule Associated Proteins (Two Groups)
Those that stabilize microtubules (Ex. Tau and EB1)
Those that destabilize microtubules (Ex. catastrophin)
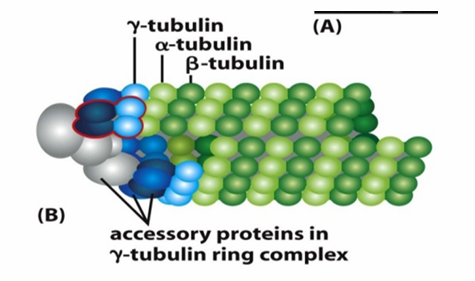
Microtubule Nucleation
Starting off growth
Involves γ‐tubulin which is present in smaller amounts in the cell compared to alpha/beta tubulin
Helps form γ‐tubulin ring complex (γ-TuRC)
Nucleates at (-) end of a new microtubule
Forms a template for the growing (+) end
γ-TuRC acts as a cap of the (-) end while microtubule growth occurs at (+) end
MTOC (Microtubule Organizing Center)
A specific location inside the cell where microtubule nucleation occurs
In animal cells, the MTOC is centrosome (red dot)
Located near nucleus
MTOC: Centrosome and yTuRC
Consists of 2 cylindrical structures called centrioles (inside centrosome which is in green)
Also has pericentriolar material (PCM) containing many γ‐TuRC complexes (red rings on green ball)
(-) end of microtubules are nucleated at the γ‐TuRC
(+) end are directed towards the cell periphery (shown as +)
MTOC role in Mitosis
The MTOC (centrosome) organizes microtubules that form the mitotic spindle.
The spindle’s microtubules attach to chromosomes to separate replicated sister chromatids.
Centrosomes are duplicated before mitosis, creating two MTOCs that move apart to opposite poles.
Microtubules nucleate from the γ-TuRC complexes at each MTOC, with plus ends growing outward.
Microtubule Toxins: Cholchicine
Useful in lab to arrest the cell cycle
Ex. cholchicine
Derived from meadow saffron
Inhibits polymerization
Binds and stabilizes αβ‐tubulin dimers
Prevents addition/loss of tubulin dimers
Arrests cells in metaphase without chromatid seperation
Microtubule Toxins: Taxol
Useful in lab to arrest the cell cycle
Taxol Function
Binds to β‐tubulin to increase affinity for (+) end
Prevents depolymerization
Prevents assembly of mitotic spindle to inhibit mitosis
Used in cancer treatment
Hard to synthesize in lab so it’s derived from pacific yew tree
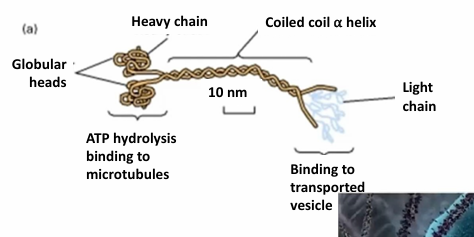
Kinesin Motor Protein
(+) directed transport on microtubules, so towards cell periphery away from MTOC
Tetrameric complex made of 2 heavy chains and 2 light chains
The globular heads (motor domains) cyclically bind to microtubules
Generates movement through ATP hydrolysis
The tails determine specificity of cargo binding
The tails are highly variable
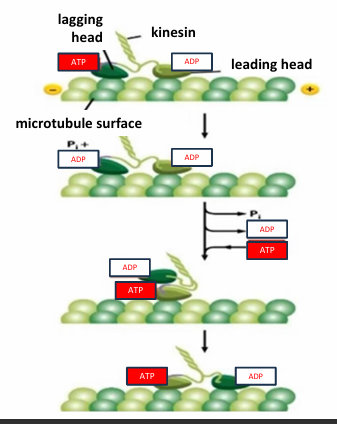
Kinesin Mechanochemical Cycle
The lagging head is bound to ATP
The leading head is bound to ADP
ATP kinesin has a higher affinity for the microtubule than ADP bound kinesin
The ATPase motor lagging head hydrolyzes ATP to ADP + Pi
Reduces affinity of lagging head for microtubule
ADP is exchanged for ATP in leading head
Increases affinity of leading head
The binding of ATP induces conformational change causing lagging head to swing in front to another microtubule binding site
This resets cycle to the top
How does kinesin move along microtubules?
Kinesin moves in a “hand-over-hand” fashion.
It has two motor heads (domains), and one is always attached to the microtubule.
The two heads work in a coordinated cycle, each in a complementary stage of ATP binding or hydrolysis.
In-vitro assays for kinesin movement
Nomarski Microscope
Following plastic beads tethered to kinesin
The track is anchored to the dick made from purified tubulin
Gliding mobility assay
kinesin are tethered to a glass slide at their cargo (Tail) ends
They can then move fluorescently labeled microtubules added to solution above slide
Dynein
(-) directed, moving towards MTOC
2 main forms: Cytoplasmic and Axonemal
Has 2 heavy chains and a variety of intermediate and light chains
Two forms of dynein
Cytoplasmic
Associated with microtubules
Direct movement of organelles and vesicles in cytoplasm
Axonemal
Found in structures powering movement of whole cells
Ex. cilia or flagella
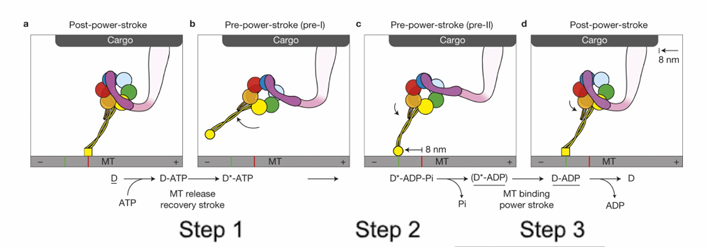
How does dynein move cargo along microtubules?
Movement is powered by a power stroke in the linker arm (near the cargo attachment site).
In a dynein dimer, the two motor units alternate power strokes, producing continuous movement.
Describe the steps of dynein’s ATP-driven power stroke
ATP binding releases motor head group from microtubule
ATP hydrolysis creates dynein-ADP+Pi that can now attach to the microtubule
The release of Pi powers the power-stroke of the liner
Pulls the cargo
Each power stroke, the cargo moves towards the (-) end by 8mm
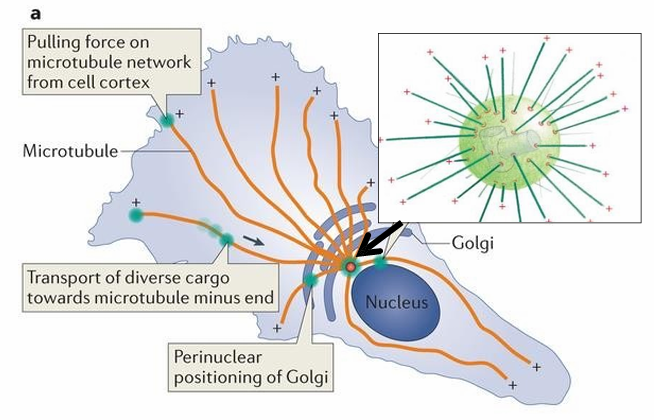
Bidirectional Vesicle Movement: Neural Cells
Microtubules span the axons of neural cells
The (-) ends are anchored to MTOC
The (+) ends extend along the axons towards synapse cell membrane
Vesicles with NTs are carried from cell body to synapse along microtubules
Microtubule Tug of War
Model describing the movement of proteins if they’re bidirectionally transported
The final direction of movement is the winner of this ‘battle’
There are regulatory proteins controlling direction in response to cell signals
Change of Direction (Microtubule Transport) Application: Melanosomes in Fish
Melanosomes: Pigment-filled organelles
Movement of it changes skin cells in response to behavioural signalling
This movement is done by molecular motors carrying it to the cell periphery or center
Dynein: Move towards (-) end MTOC
Kinesin: Move towards cell periphery (+) end
Dispersion to periphery = cell appears darker
Concentrated in middle = Cell appears lighter
This is controlled by signals using cAMP as a secondary messenger
What are the 3 types of filaments making up the cytoskeleton
actin
microtubules
intermediate filaments
Function of Cytoskeleton
Provides shape and structure
Responsible for the specialized structures in cells
Microtubules in cilia
Actin filaments in imcrovilli
The shape depends on functions
What is the dynamic nature of cytoskeleton important for
Cells that move
Cells that undergo migration or cell division
What are the three types of fibres in eukaryotic cells defined by (cytoskeleton)
Diameter
Type of subunit used to build the filament
Actin Filament Labelling (2)
Labeled using fluorescently-tagged phalloidin
Toxin derived from death cap mushroom
Binds to actin monomers with high affinity and specificity
Stabilizes the filmament when bound
Labeled with antibody
Labeled with protein fusion (Actin:GFP)
Intermediate filament labeling
Labeled using an antibody specific to a monomeric subunit
Labeled using GFP-fusion
Microtubule labeling
Labeled using antibodies specific to one of the tubulin subunits
Labeled using protein fusion (Tubulin:GFP)
Actin composition
Thinnest filament
5-9nm
Made of 2 strands of helical polymers that spiral around eachother
Each strand is built from single actin monomers
G-actin
Microtubule Composition
Thickest fibres
Made of dimeric subunits of alpha and beta-tubulin
Intermediate filament (IF) composition
There are many types
Each is assembled from a different protein or set of proteins
Epithelial Cell 3 cytoskeletal fibre Distribution
Actin (red) forms the shape of the microvilli at apical side of cell surface
IF (blue) span to provide structural support
Microtubules (green) form networks for transport
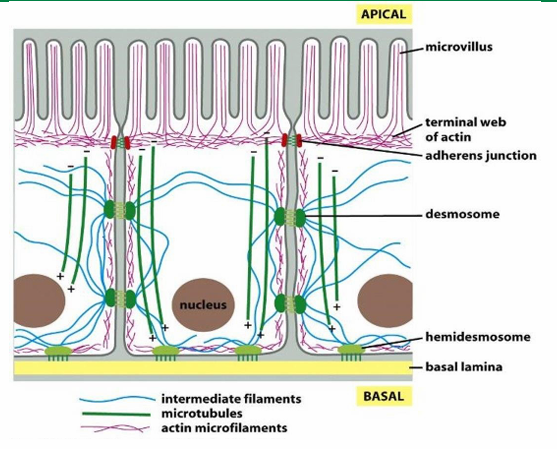
Filament-Specific Motor Proteins
Move along the actin and microtubules
No motor proteins found for IF
Which motor protein moves along actin filament
Myosin
Which motor proteins move along microtubules
Kinesin
Dynein
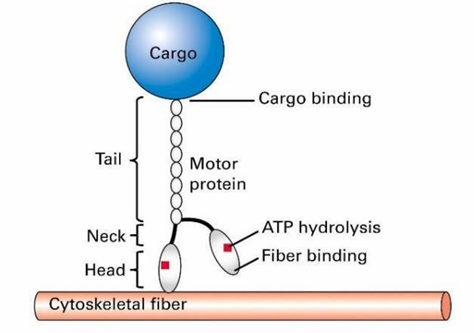
General Structure of Motor Proteins
They step along their respective fibres using cycling chemical reactions
The head domains bind to a cytoskeletal fibre
Tail domain attaches to cargo
ATP hydrolysis provides energy for this movement
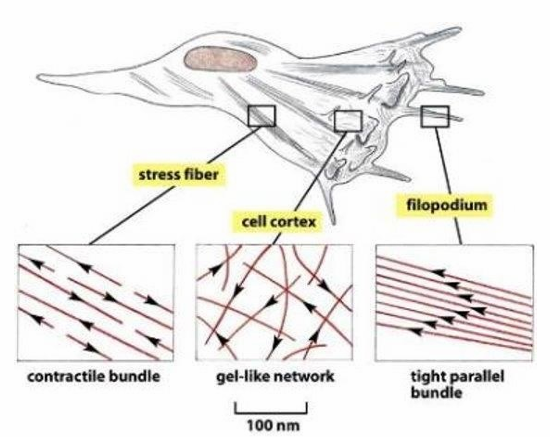
Actin-Based Structures
Highest density of actin is at cell periphery to determine shape and movement of cell surface
Establishment of microvilli
Formation of contractile bundles forming sarcomeres
Contractile ring directing cytokinesis
Actin filament organization variance within a single cell
Contractile Stress fibres (seen throughout)
Gel-like network (seen at cell cortex)
tight parallel bundles (seen in filopodia)
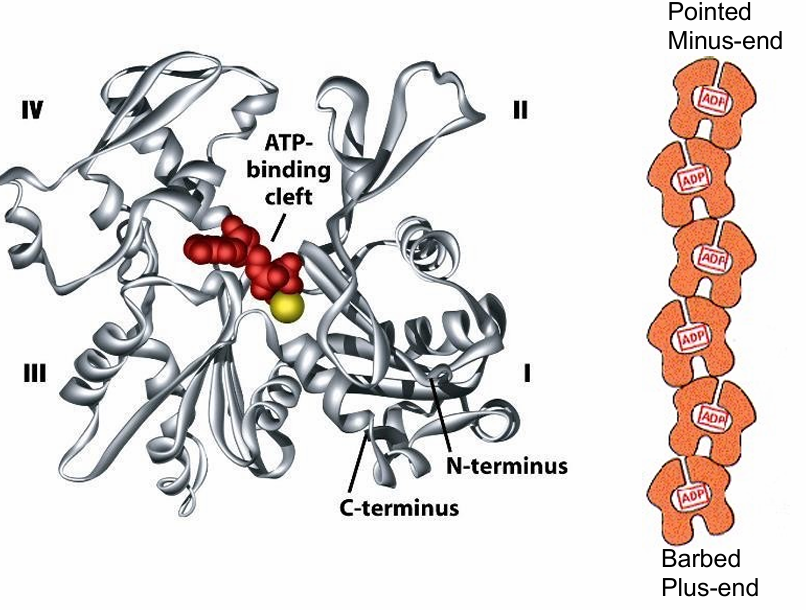
Actin Filament Polarity
No visible without the myosin proteins
They bind to actin in one orientation, pointing away
This defines (+) / (-) end of the filament based on rate of actin polymerization
(+) grows more quick and has barbed appearance
(-) end grows slower, or may shrink and has pointed appearance
G-actin structure
Has 4 structural domains
Large cleft between domains 2/4
The cleft forms ATP-nucleotide binding site
This binding site is pointed towards the minus-end
Makes them hidden as the monomers bind
Only 2 monomers at the end have exposed sites
Each actin monomer is polar so the microfilament is polar
Actin Dynamic Polymerization
Depolymerization and polymerization can occur at both the plus and minus ends
More growth tends to occur at (+) while there’s shrinkage at (-)
This is bc of ATP
When monomers are bound to ATP, they can join
Intrinsic ATPase activity hydrolyes ATP to ADP
ADP never gets released as the binding site in covered
(+): Actin-ATP monomers are added
(-): Actin-ADP comes off
Cytosol: Free actin-ADP exchanges ADP for ATP
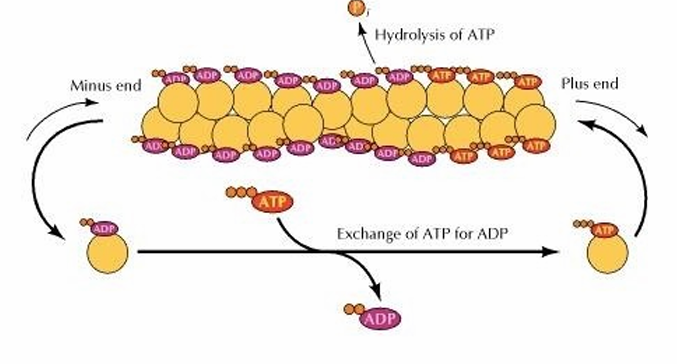
Critical Concentration
Concentration where the rate of actin monomer addition is equal to the rate of removal
No net growth at that end
If [monomer] exceeds this, polymerization exceeds rate of depolymerization (filament grows)
If [monomer] is lower, depolymerization exceeds (filament shrinks)
The critical and working concentrations are different at each end
Proteins involved in actin polymerization/depolymerization
Profilin binds to actin-ATP
Activates monomer
Promotes ATP binding
Profilin-actin dimers accumulate at plus end
Increases [monomer] at that end
Thymosin binds to actin monomers
inhibits polymerization
Thymosin-actin dimers accumulate at plus end
Creates a buffer of stored actin monomers
Treadmilling
When there is no net increase in actin filament length
Happens when rate of polymerization at (+) = depolymerization at (-)
The relative position of the filament changes to move forward
Helpful for cell movement/migration
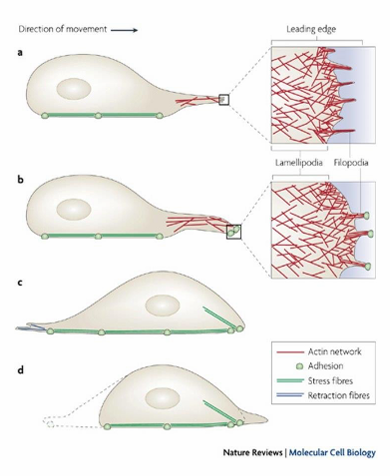
Actin Filaments and Cell Migration
Powers cell movements through organization of actin filaments to push out cell membrane
Observed through formation of filopodia and lamellipodia in a migrating cell
Forms leading edge of cell
Forms fan-like expansions of cell membrane (lamellipodia)
Forms finger-like filopodia extensions of cell membrane
Initiates movement to desired direction
Myosin Motor Protein Types
Power intracellular cargo trafficking
Myosin I / II / V are in all euk cells
Have motor domain (head) at N-terminus
Binds actin filaments
Hydrolyzes ATP to drive motor
Have different tail domains
Carries cargo at different rates
They usually move toward the (+) end of actin
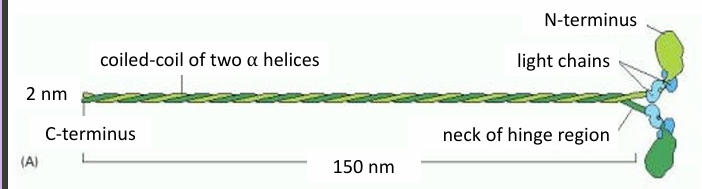
Myosin II Structure
Has 2 heavy chains forming a coiled-coil motif (green)
Has four light chains (blue)
Myosin II Mechanism
MLCK (myosin light chain kinase) phosphorylizes myosin light chains
Drives polymerization of myosin by
initiating extension of their tails
activating actin-binding domains on head
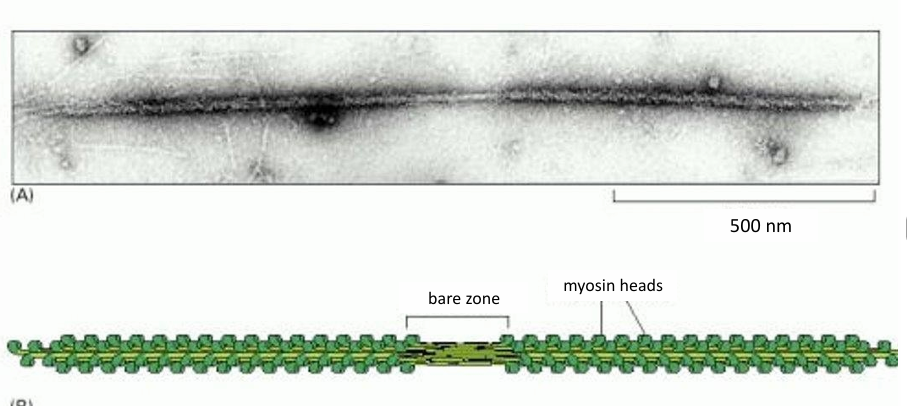
15-20 myosin II form a bipolar filament
Myosin II thick filament
Myosin ll doesn’t carry cargo
Generates contractile forces needed for many cellular activities
Myosin II Bipolar Thick Filament
Has myosin motor heads on both sides of a bare patch (zone of myosin tails)
Motor heads are exposed to be associated with actin filaments
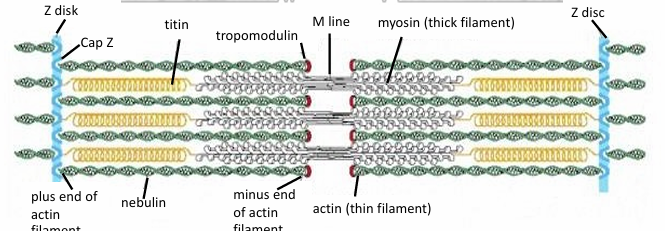
Myosin II Function in Skeletal Muscle Fibers
Sarcomeres: Structure where myosin II thick filaments associated with thin actin filaments
(+) of actin are fixed to Z-discs within the sarcomere
Between parallel actin fibers, myosin thick filaments are present
They’re also attached to Z-discs, but with titin
It’s a giant molecular spring
During muscle contraction, myosin thick filaments interact with actin to move the Z lines closer together
Muscle Contraction Mechanism
Myosin heads associate with actin filaments
They get pulled past myosin toward the middle
Occurs by the cyclical association of actin filaments with myosin motor heads
Myosin head cycles through ATP binding and hydrolysis
Allows it to move along actin filaments
Moves towards (+) end
Causes sarcomere shortening without changing any filament length
The process is calcium-dependent
Calcium Dependence of Muscle Contraction
Allows for exposure of myosin binding sites along actin filaments
After contraction Ca++ dissociates from actin filaments
Myosin heads then release the actin
The filaments slide past eachother to allow for muscle relaxation
Muscle Contraction: Chemical and Mechanical Energy
Muscle contraction involves converting chemical energy into mechanical
This is mediated by myosin
It undergoes a series of conformation changes (Mechanical) regulated by ATP binding/hydrolysis (chemical)
The steps of both cycles are interlinked to form the myosin cycle
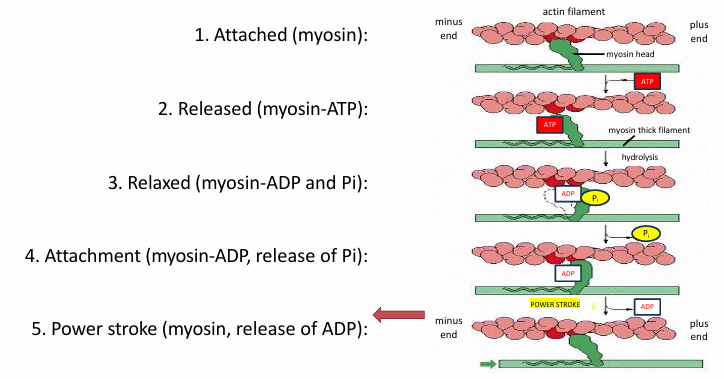
Cycle for Single Myosin Motor Head
Myosin is attached to actin
ATP binding to myosin releases actin
ATP is hydrolyzed into ADP and Pi by myosin head
Changes myosin conformation returning it to relaxed
Release of Pi increases affinity of myosin head for actin
Allows binding
Release of ADP from myosin head changes conformation
Since myosin is attached to actin, it pulls the filament
Puts cycle back in step 1
ATP binding will then release myosin from actin again
This cycle repeats many times during muscle contraction
One ATP molecule binding/hydrolysis moves the myosin motor a few nm along the actin track
Myosin V
Powers intracellular trafficking of cargo along actin
Myosin V: Melanosomes
Melanosomes are membrane-enclosed organelles containing melanin in melanocytes (skin cell type)
Each melanocyte has several dendrites stretching to connect with many keratinocytes
Incorporation of melanin into the keratinocytes of skin cells and distribution of this pigment protects cell’s DNA from UV damage
Tanning
Myosin V distributes melanosomes along actin filaments
Loss of myosin V function in animals
Leads to a phenotype called the dilute phenotype
Pigments associated with fur colour are not ditributed into the fur
Resulting colour is diluted
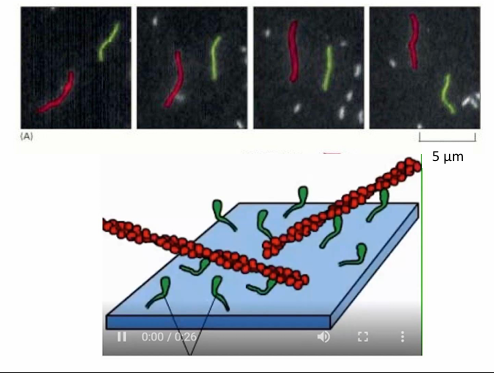
In-vitro study of myosin movement
Myosin proteins are attached by their tails to a microscope slide
Fluorescently-labelled actin filaments can be applied to the slide with addition of ATP
The chemical cycling of ATP binding, hydrolysis, ADP, and Pi power a mechanical cycle visible under microscope
Seen as movement of fluorescent actin filaments
Rates of Myosin Protein Movement
Varied with different myosin proteins
Can range from 0.2-60 micrometers/second
Rate depends on cycle of ATP nucleotide binding and hydrolysis
This varies with
Rate of ATP hydrolysis by ATPase in myosin head
The proportion of time myosin is bound to actin filament due to affinity
Myosin V spends 90% of cycle bound to actin
Myosin II spends 5%
Myosin V will move more slowly in comparison to Myosin II
Myosin Step Size
Depends on lever arm length
This is the distance by which the power stroke propels myosin
Myosin V lever is 3x longer than myosin II
Step size
Myosin II: 7nm
Myosin V: 36nm
Cargo-carrying proteins (myosin V) move in hand-over-hand fashion
Trailing myosin head detaches from actin
Gets propelled towards the (+) end of actin during power stroke of leading head
The trailing head becomes the new leading head