stupid MCB
1/110
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
111 Terms
Light Microscopy - Bright Field
Phase-contrast and DIC microscopy can be used in time lapse microscopy→ allowing the observer to study cell movement
Magnification - 0.2um
Light Microscopy - Fluroescence
localising molecules within a cell by light microscopy is fluorescent staining of cells and observation by fluorescence microscopy
Magnification - 0.2um
Light Microscopy - Intracellular ion
concentrations within live cells can be determined with ion-sensitive Fluorescent dyes
- Fluorescent dyes (e.g.. Snarf-1) that are sensitive to H+ concentrations can be used similarly to monitor cytosolic pH of live cells.
Light Microscopy - double label fluorescence
microscopy can visualise the relative distributions of two proteins. This technique involves using two different fluorescent dyes to label specific proteins, allowing the observation of their co-localization and interactions within the cell.
light microscope tagging with fluroescent proteins
allow visualisation of specific proteins in live cells
- Coding sequence of Green fluorescent protein (GFP) is fused to the coding sequence of a protein of interest–protein of interest is covalently linked to GFP. visualise GFP and therefore protein.
Confocal microscopy transmission (TEM)
1. Take significant time to scan each focal place → if a very dynamic process is being imaged - the microscope may not be able to collect images fast enough to follow dynamics 2. It illuminates each spot with intense laser- light → can bleach the fluorochrome being imaged and damage live cells by phototoxicity, thereby limiting the number of images that can be collected |
Magnification - 5 um
Scanning Electron Microscope
Electron dense
Views surfaces of specimens that are not sectioned
Magnification - 5um
Difference between indirect and direct immunofluorescence
| Direct | Indirect |
Time | Faster | Extra steps - slower |
Cost | Conjugated primary is more expensive | Secondary antibody relatively inexpensive |
Sens. | Weaker | Multiple secondaries bind to primary |
Advantages/ Disadvantages of Direct and Indirect Immunfluoresence
| Direct IF | Indirect IF |
Differences | Direct IF uses a single antibody directed against the target of interest. The primary antibody is directly conjugated to a fluorophore. | Indirect IF uses two antibodies. The primary antibody is unconjugated and a fluorophore-conjugated secondary antibody directed against the primary antibody is used for detection. |
Advantages | Faster time, less complicated. | Greater sensitivity than direct immunofluorescence. Cheaper. |
Disadvantages | Lower signal, higher cost, less flexibility, difficulties with labelling. |
Do you know the principles of FACS sorting for isolating cells from tissues?
Flow cytometry: fluorescence activated cell sorter (FACS) is used to isolate specific cell types.
Requires fluorescent tag for desired cell.
Processes up to 10 million cells/h
Provides cells for subsequent culturing and propagation.
Can you explain how we can isolate subcellular organelles from tissue or cell cultures? How do we prove that we've been successful in the isolation?
FACS: isolate specific cell types
Subcellular fraction: the study of individual compartments.
DIFFERENTIAL CENTRIFUGATION:
- Different organelles are separated at different speeds based on their size and density. Nuclei at lower speeds, mitochondria at medium speeds, ribosomal subunits at higher speeds as well as cytosol.
DENSITY GRADIENT CENTRIFUGATION:
- Organelles of similar mass are separated by centrifugation of a gradient solution according to their equilibrium density.
Organelle purity assessment:
1. Marker enzymes: measure activities of enzymes that are only found in a particular organelle.
Organelle and marker:
Mitochondrion - Cyt c oxidase, peroxisome - catalase, nucleus - RNA polymerase
2. EM: to visualise the morphology of specific organelles.
3. TEM assesses organelle purity.
CELL TO CELL ADHESION:
Occurs through specialised membrane proteins called cell adhesion molecules (CAMS).
CELL TO MATRIX ADHESION:
Occurs through adhesion receptors binding to components of the extracellular matrix (ECM)
Types of Junctions
Junctions | Adhesion type | CAMS of adhesion receptors | Cytoskeletal attachments | Intracellular adapters | Functions |
Anchoring Junctions: 1 - Adherens | Cell to cell | cadherins | Actin filaments | Catenins, vinculin | Shape, tension, signalling, force transmission |
2 - Desmosomes | Cell to cell | Desmosomal cadherins | Intermediate filaments | Plakoglobin, plakophilin, desmoplakin | Strength, durability, signalling |
3 - hemidesmosomes | cell-matrix | Integrins | Intermediate filaments | Plectin, dystorin/BPAG | Shape, rigidity, signalling |
4 - focal, fibrillar + 3D adhesions | cell-matrix | Integrins | Actin filaments | Talin, kindlin, paxillin, vinculin kinase | Shape, signalling, force transmission, cell movement |
How do cells adhere to the extracellular matrix?
Cell to matrix - Adhesion molecules - often referred to as adhesion receptors , reason why we call these receptors is because there's often ligands or things that bind to the receptors in the extracellular matrix
The cells adhesion molecules that will bind the cells together they are attached to some kind of linker molecule - and that’s usually attached to some type of cytoskeletal feature
Can you list and discuss the different families of Cell Adhesion Molecules (CAMs) and Cell Adhesion Receptors? What is important about their molecular structure? Have you considered the experiments discussed in the lectures that help us understand how these molecules have their function?
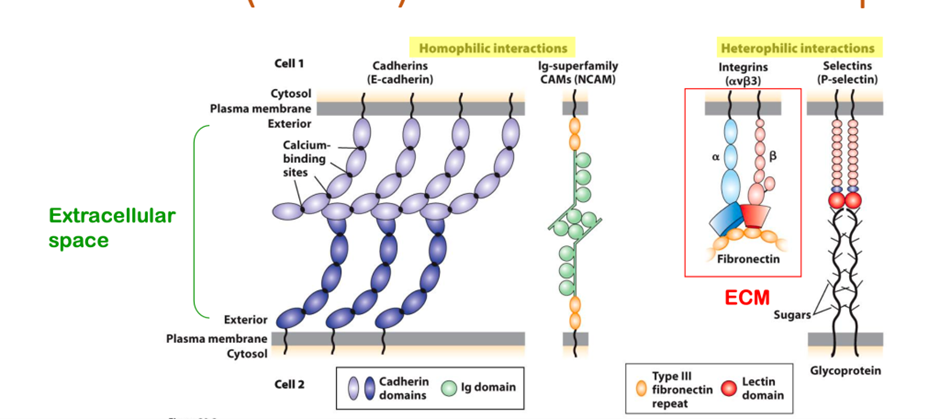
Four major families of cell-adhesion molecules and adhesion receptors that exist
1. Cadherins
2. ig superfamily
3. Integrins
4. Selectin
CAMS promote cell-cell adhesion, which involves both cis and trans binding interactions which are mutually reinforcing.
Include selectins: special example of CAMS that promote leukocyte-endothelial wall adhesion.
Calcium dependent CAMS: integrins, cadherins, selectins.
Calcium independent CAMS: Ig-superfamily.
Can you compare and contrast the different types of cell-cell junctions in terms of: 1) the structure and function of the junction; 2) the types of molecules involved; 3) the important adaptor molecules; 4) the cytoskeletal attachments in each case.
Junction | Adhesion type | CAMS/Adhesion receptor | Cytoskeletal attachments | Intracellular adapters | functions |
Tight junctions | cell-cell | Occludin, claudins, JAMs | Actin filaments | ZO-1,2,3, PAR3, cingulin | Controlling solute flow, signalling + polarity |
Gap junctions | cell-cell | Connexins, innexins, pannexins | Via adapters to other junctions | ZO-1,2,3 | Communication, small-molecule transport between cells |
Plasmodesmata | cell-cell | Undefined | Actin filaments | NETIA | Communication, molecule transport between cells. |
Cell-ECM adhesion:Hemidesmosomes
- Connects 2 cells together → interacts with the intermediate filament via adapter molecules
The actual cell adhesion receptor molecule involved = Integrin → made up of Alpha and Beta subunitis (both membrane bound units) they attach to different adapter molecules really critically
Epidermolysis
Integrins link to the cytosolic intermediate filament and basal lamina via laminin
Focal adhesions:
- Attach cells to EXM
- Involve integrin molecules
- In a focal adhesion rather than the intermediate filament seen in hemidesmosomes , Actin filament are the cytoskeletal feature of the cell that’s attaching to the ECM that are the ultimate attachment to the cell via a whole range of different proteins
Important protein adapters important for the focal adhesion to operate
- Talin
- Vinculin
- Alpha actinin
- Paxillin
- Focal adhesion kinase
- SRC
These focal adhesions are attached to the ECM via a different protein by protein called Fibronectin
- These focal are non epithelial cells
COMPONENTS OF THE EXTRACELLULAR MATRIX:
COMPONENTS OF THE EXTRACELLULAR MATRIX:
Major ECM components:
Cells (fibroblasts, mast cells, macrophages, capillary cells), water, lipids, ions, proteins (listed below)
Proteins of ECM:
1. Insoluble (structural): collagen, elastin
2. Attract water: proteoglycans
3. Soluble (multi-adhesive matrix proteins): laminin, fibronectin
Protein | Function |
Collagen | Provides structural support to ECM. Insoluble. |
Elastin | Insoluble, structural. |
Proteoglycans | Heavily glycosylated, has covalently linked polysaccharide chains called glycosaminoglycans (GAGs), negatively charged, bind water. |
Laminin | Multi-adhesive protein, forms network. Integrins react with laminin. |
fibronectin | Integrins mediate cell-ECM adhesions via fibronectin. Adhesive protein. |
Basal lamina (epithelial cells)
the foundation for assembling cells into tissues. A sheet-like network of ECM components. 60-120nm thick and synthesised by the cells that rest on it. Associates with plasma membrane of cells above and thicker collagen fibres below.
Comprised of four ubiquitous protein components:
1. Laminins: multi-adhesive protein, forms network
2. Perlecan: proteoglycan, crosslinker
3. Nidogen (entactin): crosslinker
4. Type IV collagen: trimeric molecules, forms network
Can you explain how integrins bind to fibronectin? Do you understand the experiments discussed in lectures that help with this understanding?
Integrins are transmembrane receptors that assist in cell-cell and cell-matrix adhesion. Integrins mediate cell-ECM adhesions via fibronectin. Integrin binds to fibronectin via the RDG motif.
Can you discuss the steps in leukocyte extravasation?
- White blood cell wanting to exit a blood vessel wall to reach a site of infection or inflammation
We have -
- Endothelial cells- Blood vessels
- Leukocyte
- ICAMs
- In the white blood cell we have cell adhesion receptors (aL and B2 integrin)
- Specific carbohydrates ligands on the surface of the white blood cell
Within the endothelial cell and within the vesicle there are also specific molecules (CAMs) selectin molecules, usually wrapped in a vesicle - not presented to the cell surface unless something happens
- Inflammation occurs within the tissue
- that inflammation activates a factor = PAF - this will lead P-selectin moving out of its vesicle and being presented to the external part of the endothelial cell - these P-selectin will bind a selectin ligand which starts to slow the leukocyte down (slow rolling interaction)
- This will impact the integrins and activate PAF - which will bind the receptor for for PAF on the white blood cell
So the binding of PAF to its PAF receptor will activate integrin molecule which will then attach to ICAM - and a strong interaction persists
- Now you've got integrin molecules (from the white blood cell) binding to adjacent endothelial cells , two endothelial cells will then open up and the white blood cell will be able to extravasate right into the tissue itself
○ Can you summarise the two pathways of protein sorting?
1. Pathway 1: Proteins contain signal sequence → ribosomes attach to the ER membrane and translocation is co-translation
2. Pathway 2: Proteins do not contain signal sequence → ribosomes complete translation in the cytosol and translocation is post-translation
Can you describe the sorting signals which direct post-translational or co-translational proteins to organelles?
Can you explain the translocation process for proteins to enter the nucleus, mitochondria, peroxisomes and endoplasmic reticulum?
PATHWAY 2
| Nucleus | Mitochondria |
Protein | Folded | Unfolded |
Chaperones required |
| Hsp70 + Matrix Hsp70 |
Signal | +++ Either: - Continuous sequence of +ve a.a. - Bipartite sequence of +ve a.a. - 2 different distinct parts that fold to be close together | - Most contain Mitochondrial Targeting Sequences (MTS) - Different signals to target proteins to different sub-mitochondrial compartments
|
Cytosolic Receptor | Importin | N/A |
Membrane Receptor |
|
|
Translocon ** protein complex that mediates protein transport across cellular membranes | Nuclear Pore FG Nucleoporins - Nuclear Pore is composed of 8 structural subunits surrounding a central channel - FG nucleoporins line to NPC channel and form a hydrophobic gel-like matrix that regulates diffusion/transport | TOM and TIM components - Both rich in acidic -ve a.a. - Outer membrane - TOM 20/22 (import receptor), TOM40 (general import pore) - Intermembrane space - TIM23/17/44 |
Energy | Ran protein (binds to importin) | ATP (chaperone function), proton motor force (H+ gradient) |
Cleavage | No | Yes Path A No Path B
|
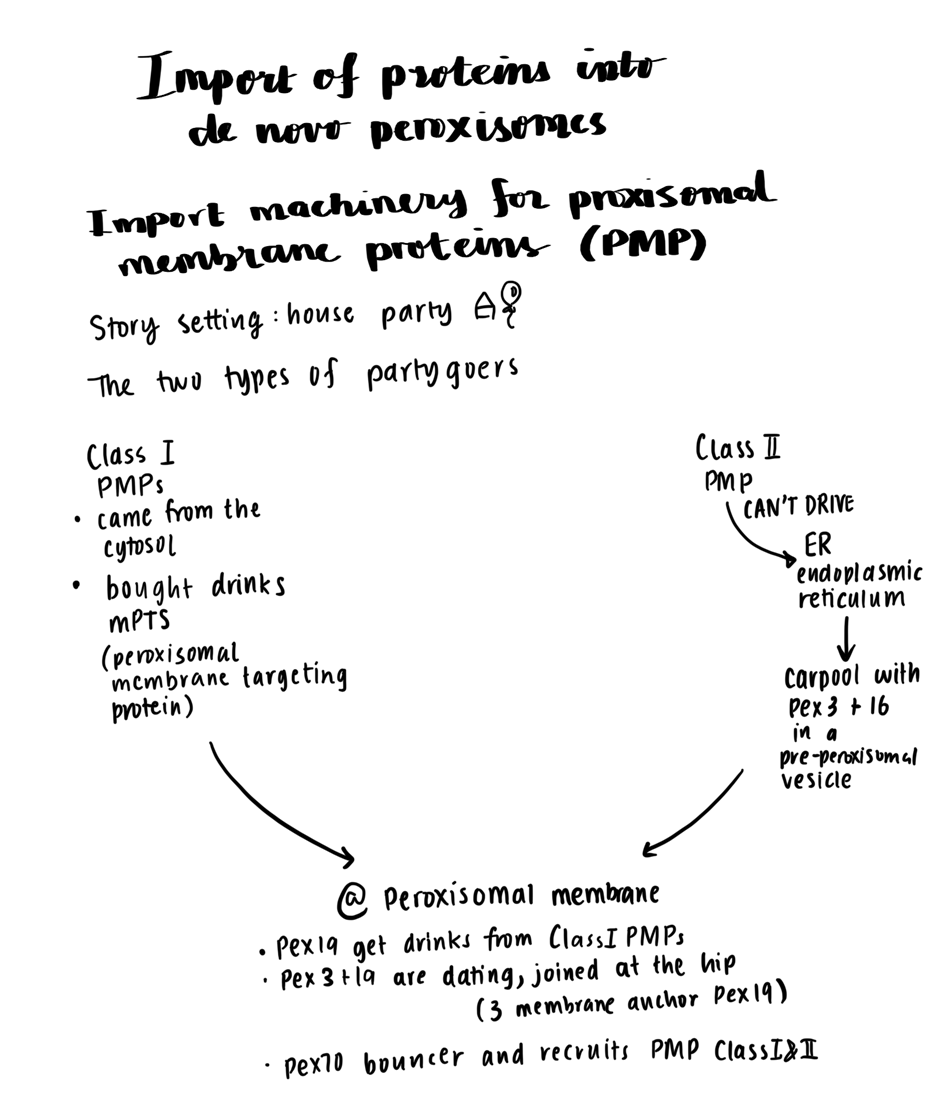
| Peroxisome | |
Protein | Folded | |
Chaperones required | - | |
Signal | PTS1 signal: signal at the extreme C-terminus PTS2: signal near the N-terminus | |
Cytosolic Receptor | PTS1: Pex5 PTS2: Pex7 dependent, interactions with Pex5
| |
Membrane Receptor |
| |
Translocon ** protein complex that mediates protein transport across cellular membranes | PTS1 - Pex13/14 complex - Pex13 has YG repeats (similar to FG repeats of the FG nucleoporins) | |
Energy | ATP for Pex1/Pex6 | |
Cleavage | PTS1 - No PTS2 - Yes |
IMPORT OF PROTEINS INTO DE NOVO PEROXISOMES
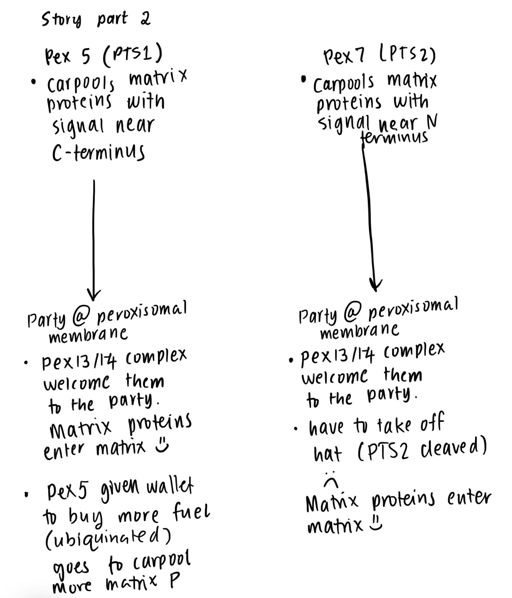
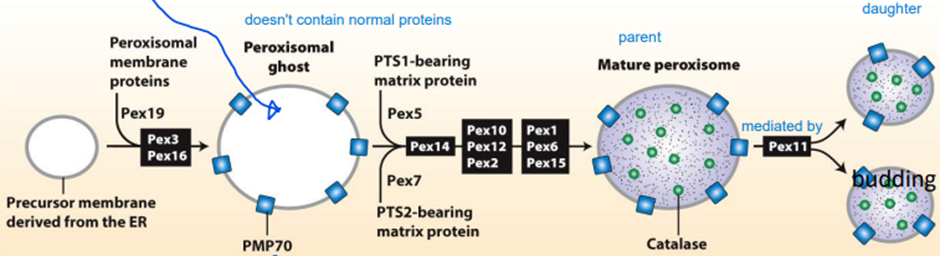
PATHWAY 1
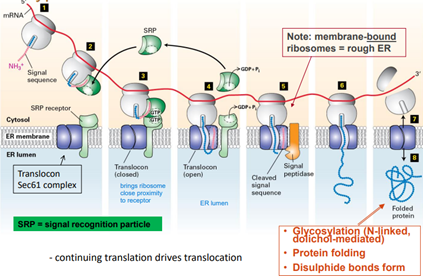
ENDOPLASMIC RETICULUM
Signal sequences
- 16 - 30 amino acids
- one/more +ve a.as near N terminus
- 6 - 12 hydrophobic residues that bind the signal recognition particle (SRP)
- The signal peptide is cleaved from pre-cursor protein\
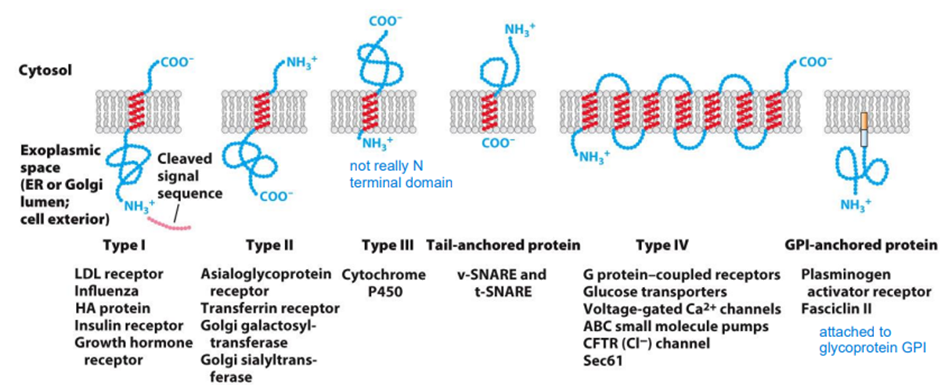
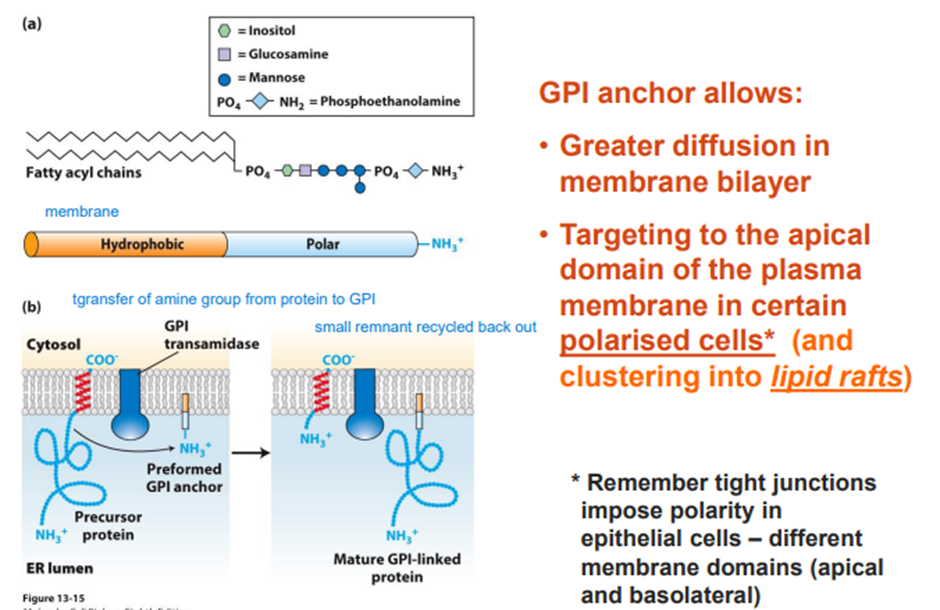
○ Identify the principal components of the secretory pathway.
○ Outline the final destination of proteins leaving the Golgi.
○ Describe the protein modifications that occur in the endoplasmic reticulum.
1. Proteolytic cleavage
a. Removal of signal sequence by signal peptidase
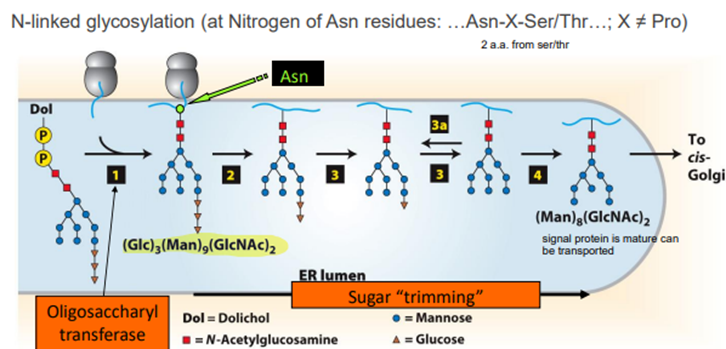
2. Glycosylation
a. N-linked glycosylation
Dol transfers an oligosaccaride to a basic a.a. On a synthesising protein, The glucose is removed leaving the mannose and n-acetyl glucosamine moiety.
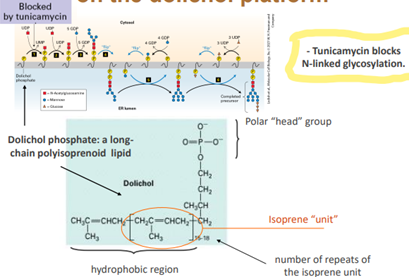
N-linked glycosylation is blocked by tunicamycin
3. Disulphide bond formation (+ rearrangement)
- Catalysed by protein disulphide isomerase (PDI)
4. Protein folding and oligomerisation
- Require molecular chaperones and other ER proteins
- BiP and lectins (calreticulin and calnexin) act as chaperones to prevent incorrect folding
- PDI enhances correct folding through creating correct disulphide bonds
Unfolded Protein Response (UPR)
- Induced by the accumulation of unfolded proteins in the ER and tunicamycin
- Results in the activation of Hac1 (XBP1) transcription factors. Hac1 increases the expression of proteins that promote protein folding.
- Increased production of pro-folding chaperones
** retained proteins undergo mannose trimming. Man5-6(GlcNac)2 recognised by OS9 → dislocated across the ER membrane to the cytosol for degradation.
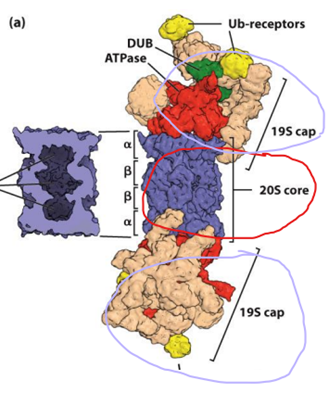
- Ubiquitin ligase adds 4 ubiquitin to the dislocated proteins as they enter the cytosol. These polyubiquinated proteins are degraded by the 26S proteasome. The 26S proteasome is comprised of a 20s proteasome (catalytic core) with 19S proteasome caps
What is the difference between anterograde and retrograde transport?
Anterograde transport = towards the plasma membrane whereas retrograde transport = back towards the nucleus/ER
Retrograde targeting signals:
- ER lumenal proteins: …KDEL-COOH
- ER membrane proteins: …KKXX-COOH
Golgi processing
- O-linked glycosylation
- Carbohydrate chains attached to the hydroxyl (-OH) group of serine and threonine residues
- 1 - 4 linear sugar residues
- Complex N-linked glycosylation
- Carbohydrate chains attached to the amide nitrogen of asparagine residues
- Several longer branches of sugar residues
○ Explain cisternal maturation of the Golgi complex.
○ Cisteral maturation: the mechanism in which forward transport of cargo occurs. It is when the cargo stays still but the resident enzymes are re-distributed via retrograde transport.
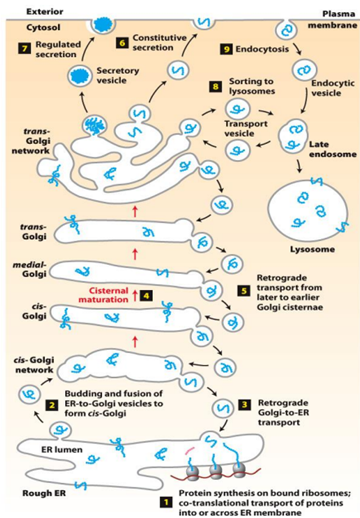
○ Demonstrate how proteins can be retrieved if mis-sorted.
○ Identify the principal components of the secretory pathway.
○ Outline the final destination of proteins leaving the Golgi.
○ No targeting sequence:
■ Soluble proteins get secreted from the cell
■ Membrane proteins delivered to the plasma membrane via bulk flow (default pathway = constitutive secretion → unregulated)
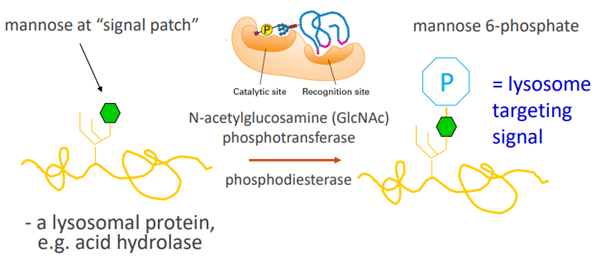
○ Targeting signal:
■ To the lysosome (M6P signal)
■ Golgi proteins have carboxyterminal “retention” signals (and are recovered by retrograde transport).
○ Describe the protein modifications that occur in the endoplasmic reticulum.
○ Explain cisternal maturation of the Golgi complex.
○ Summarise vesicle transport within the secretory pathway, including pinching off and vesicle fusion.

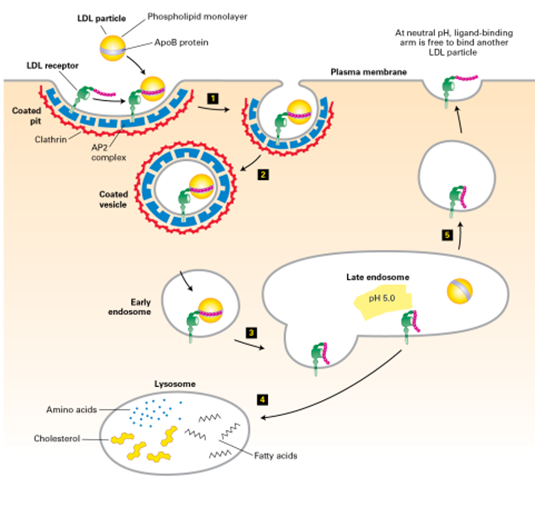
Types of Vesicles
Type of Vesicle | Where used |
Clathrin-coated - 3 heavy + 3 light chains - Has intrinsic curvature due to bend in heavy chains
| - trans -Golgi to endosome - Endocytosis (plasma membrane → endosome) |
COPI-coated vesicles | Retrograde transport - Golgi (trans-medial-cis) - Cis-Golgi to rER |
COPII-coated vesicles | Anterograde transport - ER to cis-Golgi |
Vesicle Budding
- Initiated by recruitment of small GTP-binding protein to patch of “donor” membrane that contains membrane cargo proteins
- GTP-binding protein regulates coat assembly
- Coat proteins bind cytoplasmic face of membrane (include adapter proteins which first bind the cytosolic face of membrane proteins)
- SNARE proteins are involved in membrane fusion
- Dynamin polymerises around the “neck” of the budding vesicle. Hydrolysis of GTP by Dynamin results in release of the vesicle from the donor membrane
Secretory Pathway
Know the structures, dynamics and functions of the actin based cytoskeletal system
Structures:
Actin has 2 forms: globular (G-actin) & filamentous (F-actin)
F-actin: fast growing barbed end (+), slow growing pointed end (-)
Formed by 3x G-axtin
Forms branched networks in lamellipodia and parallel bundles in filopodia and stress fibres
Dynamics:
Actin filaments undo constant assembly and disassembly = treadmilling
Dynamic turnover regulated by actin binding proteins that promote nucleation, capping, severing and crosslinking of filaments.
Functions:
Generates forces for cell motility
Facilitates intracellular transport of vesicles and organelles along actin tracks
Cofilin - Active Binding Protein
Fragments ADP-actin filament regions (-ve pointed end)
Enhances overall depolymerisation by making more filament at the -ve ends
Releases ADP-actin monomers
Severs filaments at the negative end
Generates fragments that will further dissemble
Provides G-actin for assembly at the positive end
Profilin - Active Binding Protein
Promotes polymerisation
Binds to ADP-G-actin - causes opening of the cleft increasing release of ADP and replace with ATP
Increases the local supply of ATP-G-actin
ATP-G-actin-profilin can bind to the +ve end of F-actin - profilin is released upon this interaction
Does not increase treadmilling rate, but ensures all free G-actin has bound to ATP - assisting in the increase of treadmilling
Ensures free G-actin is bound to ATP
Enhances polymerisation (can only bind to this end)
Gelsolin
Caps and severs
Regulated by Ca2+ ion concentration
Binding to Ca2+ = conformational change -> allows to bind to the side of actin filament
Gelsolin inserts itself between the subunits -> breaks the filaments
Caps the (+) end of the break preventing further assembly
Generation of multiple (-) ends from the same original filaments promotes disassembly => complete disassembly
CapZ
Capping protein
Tropomodulin
Capping protein
Arp2/3
Produces branched networks -> lamellipodium/leading edge
Requires a nucleation promoting factor (NPF - such as WASp) and a side of existing actin filament
WASp
NPF required for Arp2/3 |
Intramolecular interactions keep WASp in an inactive form WASp becomes active when its RBD binds active CDC42 and the basic (B) domain binds to the membrane lipid PI(4,5)P2 - result in exposure of A/C/W domains (???) |
Thymosin-B4
Actin monomer sequestering protein
Formin
Long actin filaments -> stress fibres
Produce long actin filament
G-actin monomers
divided into 2 subunits by ATP-binding cleft
-> -ve end: binding cleft exposed (pointed end) vs. +ve end: ATP binding cleft contact neighbouring subunit (barbed end)
-> formation of helical structures = F-actin
Polymerisation/depolymerisation rates
- ATP-G-actin binds to (+) end -> ATP eventually hydrolyzed to ADP + Pi
- Conformational change -> filament = asymmetric & responsible for different association/dissociation rates at each end
- As Pi slowly released from subunits, there are 3 distinct regions in the filament
Actin-nucleating proteins
- These can rapidly and locally nucleate actin assembly under the control of signal transduction pathways
Regulation of actin assemble
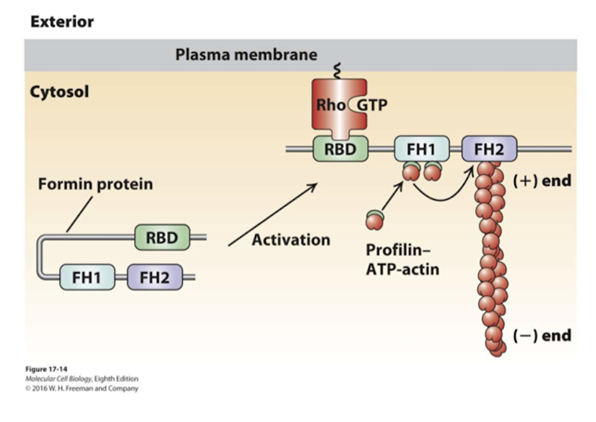
- Formins
- FH2 domains form a dimer - binds to 2 actin monomers
- Rocking back and forth - allows addition of actin monomers
- FH2 sits at the (+) end - protect it from getting capped
Rho-GTP activates Formin
- When Rho-binding domain (RBD) binds to the Rho-GTP - this allows the exposure of the FH2 domain
- F1 will bind to profilin ATP-G-actin -> accelerating assembly
Actin + endocytosis
- Actin polymerisation powers endocytic vesicles into the cytoplasm
Listeria
- Intracellular bacterium
- Intra and inter cellular movement requires actin assemble
- Actin comets
- Required for listeria motility
- ATP-G-actin
- Arp2/3 complex
- CapZ
- Cofilin
Actin-myosin interactions
- Actin drive mobility through interaction with myosin
- Myosins can convert the energy released by ATP hydrolysis to mechanical work
- Myosins - large family of motor proteins that can move along actin filaments
Class I | Single heavy chain Variable number of light chains Move towards (+) end |
Class II | Assemble into bipolar filaments containing two heavy chains Only class involved in contractile functions Short neck Two light chains per heavy chain Move towards (+) end |
Class V | Motor with two heavy chains Long neck region with 6 light chains each Tail terminates in domains which bind specific organelles Move towards (+) end |
Which end of the actin microfilament, the + or - end, has the nucleotide (ATP) binding site exposed?
Negative end has ATP cleft exposed
Positive end = back of molecule
What are the three phases of actin polymerisation?
Nucleation: Very slow. Initial pool of G-actin monomers. Formation of a nucleus comprising 3 G-actin (monomers) - rate limiting step (can be skipped if we have a nuclei)
Elongation: Diminishing pool of G-actin monomers. Actin fibrs elongate steadily in the presence of ATP/ADP bound monomers
Steady-state: polymers in ‘steady state’ with equilibrium concentration of G-actin monomers.
What is the ‘steady state’?
Equilibrium of growth -> steady amount of G-actin monomers. Neither increasing not decreasing it’s remaining steady
What is meant by ‘actin treadmilling’?
At steady state -> the (+) end is at a greater concentration than the (-) -> (+) end receiving an addition as the (-) is at a loss
Powered by ATP hydrolysis
Regulated by several actin-binding polymers
Basically, the movement in which the (+) end of the actin gains an ATP-G-actin and extends at the (+) end but loses an ADP-G-actin at the (-) creating a shortening.
What effect would you observe on actin filaments if the G-ATP-actin concentration was increased from 0.3 to 0.7uM?
Concentration above both ends = polymerisation
Concentration below both ends = depolymerisation
Actin filament dynamics can be regulated by actin binding proteins. What proteins:
- Sequester ATP-G-actin? Thymosin-B4
- Cap actin? CapZ & Gelsolin (+ end), Tropomodulin (- end)
- Nucleate F-actin? Formin, Arp2/3
- Cause actin filaments to branch? NPFs, Arp2/3 (NPFs are involved because Arp2/3 can’t do it by itself - should give both in answer)
- Fragment actin filaments? Cofilin (can only bind where there’s an ATP -> -ve end) and Gelsolin (can bind anywhere)
- Cause replacement of ADP with ATP in G-actin monomers? Profilin
- Caps AND severs actin filaments? Gelsolin
- Crosslinks actin filaments? Filamin, Fimbrin, Spectrin and a-actinin
How does actin function in endocytosis?
Arp2/3 needs an NPF for endocytosis - uses WASp. Actin will be coupled to the growing endosome and that will generate force to draw the endosome in and deliver where it needs to be - pushing on cell plasma membrane
How does Listeria hijack the actin cytoskeleton:
Listeria uses Arp2/3 to generate new actin sitting at the positive end and pushes along as the actin grows - has it’s own NPF (like WASp) on it’s surface - ActA (Acts as A NPF) - promotes Arp2/3 and assist in pushing the microorganism along.
We also need cofilin to promote severing of the negative end - make more available for growth at the positive end (polymerisation) -> pushes forward into branched actin.
CapZ used to cap any branches forming away from the cell that stops the growth of the branch chain -> ensures only growth that occurs is behind the bacterial cell to help push it into a certain direction.
What domains do myosins have?
Head, neck and tail - head makes contact to microfilament, neck for the movement
How does myosin utilise ATP to generate a powerstroke?
Binds ATP, head released from actin
Hydrolysis of ATP to ADP + Pi myosin head rotates in ‘cocked’ state
Myosin head binds actin filament
‘Power stroke’: release of P and i elastic energy straightens myosin; moves actin filament left
ADP released, ATP bound; head released from actin
- Binding of ATP: disengagement from actin
- Hydrolysis of ATP: cocking head and binding
- Release of Pi: power stroke
What is the difference between processive and non-processive myosin movement?
Processivity = constant contact
If processive, one of it’s head is always in contact with the cytoskeletal element
Processive = myosin V - 70% on the cytoskeletal element
Non processive - myosin II - 10% but there are MULTIPLE
Think about them as feet/walking
What is the structure of a sarcomere?
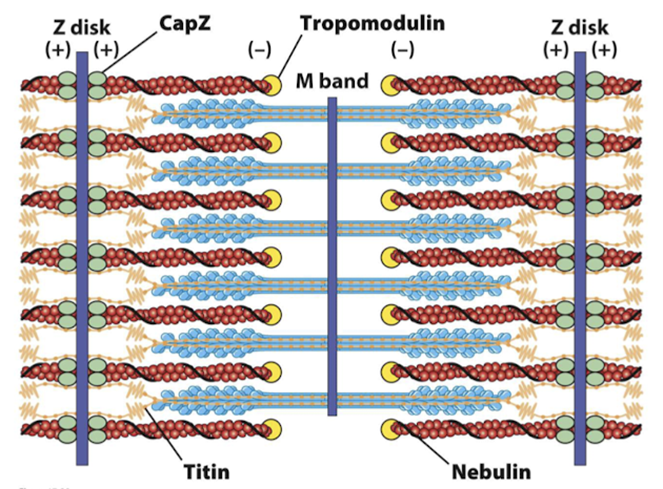
Titin - prevents overstretching and holds thick (myosin) filaments in the centre
Nebulin - determines the length of the thin (actin) filaments
CapZ & Tropomodulin - stabilise actin filament length
How do sarcomeres contract?
Contraction is regulated by calcium
Calcium is pumped in and stored within the sarcoplasmic reticulum
Tropomyosin covers the myosin binding sites on the actin filament - troponin changes shape with calcium -> causes conformational change in tropomyosin that exposes the myosin binding sites
= thin-filament regulation
Inactive state = tropomyosin
Increase in Ca -> troponin conformational change -> strips away tropomyosin -> exposes sites to contract (Ca binds to troponin)
How do smooth muscles contract?
No troponin used here
Calcium binds to calmodulin (CaM) - conformational change
CaM activates myosin regulatory LC (MLC) kinase
MLC kinase phosphorylates the MLC
Myosin unfolds and becomes active
= thick-fialment regulation
What role do focal adhesions play in cell migration?
Provide adhesion and they’re focal like dots or feet
What processes within the cell lead to protrusion of the leading edge?
Lamellipodia and filopodia that projects the membrane forward
Formation of branches actin - NOTE: be clear if microtubules or actin - what type of actin? Branched actin? Lamilopodia or filopodia? Critical for movement
What is the role of the Rho family GTPases in cells migration?
GTPases essential for formation of structures for cell migration - regulated
What kind of assay(s) could we use to measure the role of a given protein in migration?
- Scratch wound - simple, scrape cells away in middle and the other cells tend to migrate = wound closure and we can measure the rate this is happening
- If we mutate Rho-GTPases we can remove the ability to migrate together -> we can stain cells to see how actin looks
microfilaments vs microtubules
Microfilaments | Microtubules |
- ATP - 1x structure - ATP binds at (+), ADP binds at (-) - Movement: treadmilling - nucleation | - GTP - 2x subunits -> alpha and beta - y-TuRC - Generation of cytoskeleton |
Myosin V | Kinesin 1 & 2 | Dynein | |
Bind to | Actin microfilaments | microtubules | microtubules |
Nucleotide free | bound | Strongly bound (leading head) | Bound Straight linker |
ATP bound | Head released | Head bound conformation change in linker - swings forward and docks with head - swings tailing head forward (power stroke) | Head released But when minds to microtubule linker becomes bent |
ADP and Pi | Head rotates with respect to the neck “Cocked head” Binds to actin | Pi is quickly released | Pi is quickly released |
ADP | Release of Pi causes “power stroke” | Weakly bound (trailing head) | Release of Pi causes power stroke Linker straightens |
inactive | Folded - regulated by binding cargo | Folded - regulated by receptor on cargo | Requires dynactin to bind to cargo |
Know the structures, dynamics and functions of microtubule based cytoskeletal systems
Structure:
Hollow, cylindrical polymers comprised of alpha/beta-tubulin
Nucleate from MTOCs
Microtubules
- Roles in mitosis and the motile apparatus of cilia and flagella
- Involved in transport and organisation of organelles
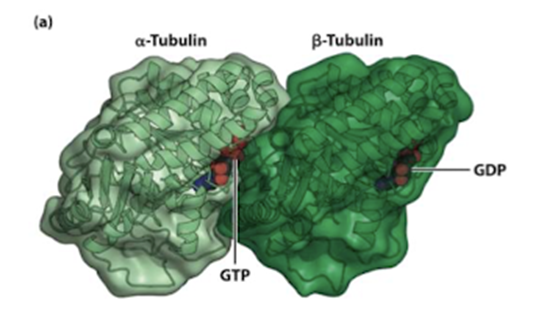
What are the subunits that construct microtubules? What nucleotides do they bind, and where are these nucleotide-binding sites?
- Dimer of alpha and beta tubular they bind thyronine nucleotides - predominantly GTP
- GDP binding clef of B is exposed by GTP of A is licked in
- Alpha cannot hydrolyse GTP but Beta can hydrolyse GDP
What is the structure of a typical microtubule?
Singlet: 13 protofilaments - alpha and beta tubule dimers. When we get a full wrap around there’s a slight angle where the seam (where they come in contact) found in cytoplasm
Doublet: 13 + 10 - found in cilia, flagella
Triplet: 13 + 10 + 10 - found in basal bodies, centrioles
Name two MTOCs found within the cell. How do they nucleate microtubules?
MTOCs: centrosomes, basal body, spindles pole, y-TuRC
They will have y-TuRC somewhere - this contains y-tubulin that is arranged to act as a template to get nucleation to occur - provides a template for the assembly of a/B tubulin dimers into the protofilament array.
What is the dynamic life of a microtubule determined by?
- Rate of growth
- Frequency of catastrophes
- Rate of depolymerisation
- Frequency of rescues
Most important thing is the catastrophes and rescues (catastrophe = ram horns, dissociating tubules)
What is the structural difference between a polymerising microtubule and a dissociating one?
GTP cap - most critical thing that determines if we have growth or disassemble or the transition from a catastrophe to rescue
Assembly - stable GTP cap - this determines what happens structurally
What is thought to drive microtubule rescue?
GTP islands introduced during repair of microtubule damage.
As long as we have a GTP cap at some point we can go into rescue by hitting a GTP island
What is microtubule capture?
Using dynamic instability to make contact with a ‘target’, with this contact stabilising the molecule.
Microtubule growing and shrinking until it comes into contact with a target - this will suppress any further catastrophe or growth - often finish temporarily because the microtubule has found what it needs - dynamic instability helps it find its target
How do colchicine and taxol affect microtubules?
- Colchicine sequesters the tubulin dimers so that microtubules cannot be assembled
- Taxol stabilises microtubules so that depolymerisation (and therefore search and capture) cannot occur. Taxol treats cells that go through mitosis that shouldn't - this is cancer cells not normal cells
In which direction do a) kinesins and b) dyneins transport their cargo?
a) All kinesins move towards the + end - except for kinesin 14 (move towards the - end)
b) Dyneins move towards the - end only
How do each of these motors bind their cargo?
- Kinesins bind directly
- Dynein requires dynactin
How does ATP hydrolysis drive movement of each of these molecules along microtubules?
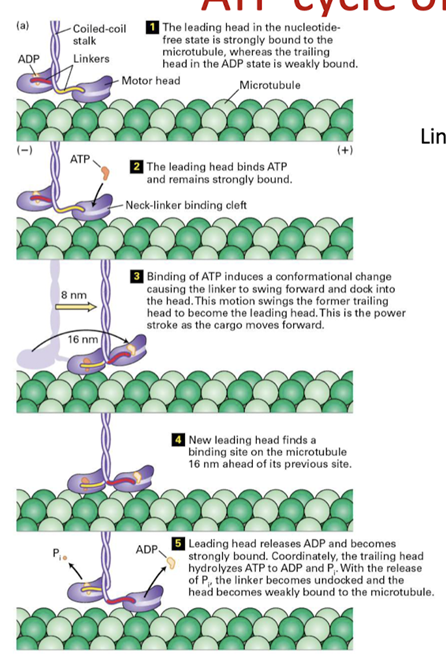
- How does ATP drive these?
- Dynein similar to myosin but kinesin very different
- Domains as functional regions - stalks that make contact it’s not the stalk the moves its the linker - what generates this movement?
- Myosin = neck region, dynein and kinesins = the linker
What is the general structure of cilia and flagella?
Nexons - interlink doublets of microtubules
How do cilia and flagella bend?
Bending - nexon resisting the sliding activity causing a bend. When dynein is switched off, bending is relieved.
What medicates the increased MT dynamics during mitosis?
XMAP215 suppresses catastrophes induced by kinesin 13 byt XMAP215 is inactivated by phosphorylation during mitosis.
Kinesin 13 is potent - usually suppressed by XMAP215 - they compete for whats happening in the subunit and dimers at positive end
Increased MT dynamics is by inactivating XMAP215 by phosphorylation = reduction activity very specifically during mitosis
Take cells, syncrhonise them to mitosis at same stage and harvest them = low levels of XMAP215 interfase = high level of XMAP215
How do sister chromatids come to be aligned at the metaphase plate?
Microtubule search and capture followed by congression that involves bidirectional movement. Multiple motors contribute to this.
Microtubule search and capture grabs them and then attachment
Backwards and forwards movement until they’re aligned - congressional rate
Whole lot of kinesis that contribute to this activity as well as natural growth and shrinkage of MT themselves
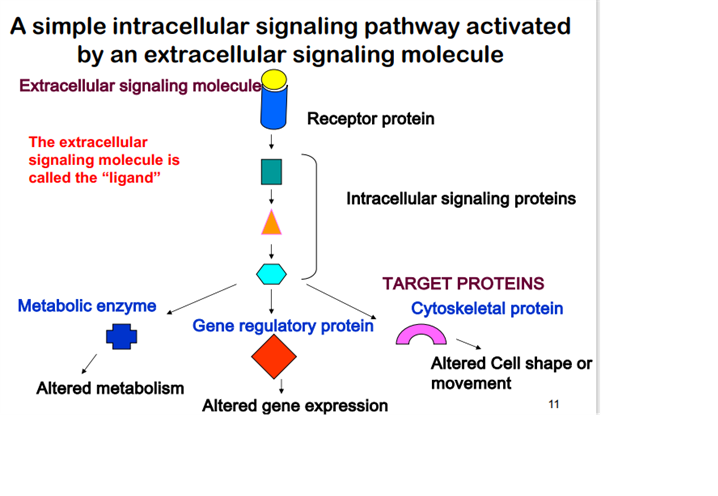
Understand the basic principles of signal of signal transduction
Intracellular signal pathway activated by an extracellular signal molecule. These signal pathways causes have individual signal proteins that result in the activation and regulation of metabolic enzymes, gene expression and cytoskeleton proteins.
Hydrophilic vs Hydrophobic signals
Hydrophobic signals do not require an membrane receptor in order to activate it’s intracellular signal proteins.
- Move through membrane without a cytosolic receptor
Hydrophilic signals
Hydrophilic signals require a membrane receptor that activate more transduction properties and second messenger.
Secondary messengers
Many signals are mediated by secondary messengers, that amplify signal.
Four features of signal transducing system
Specificity
When signal molecules binds to their complementary receptor. Non complementary signal will not bind.
Amplification
When enzymes activite enzymes, the number of signal affected molecules increase geometrically in an cascade.
Desensitization/ Adaptation
Receptor activation motivation triggers a feedback circuit that shuts off the receptor or removes it from the cell surface.
Feedback mechanisms that turns off the receptor.
Integration
When two signals have opposite effect on a metabloc characteristic such as the concentration of a second messenger X, or the membrane potential Vm the regulatory outcome results from the integrated input from both receptor.
Two major types of cellular responses
Changes in activity or function of specific pre-existing proteins
Proteins activated by a covalent change or conformational change.
Changes in amount of specific proteins
Making less or more of something to archive specific goal by modifying transcription factors.
Turning signals on and off
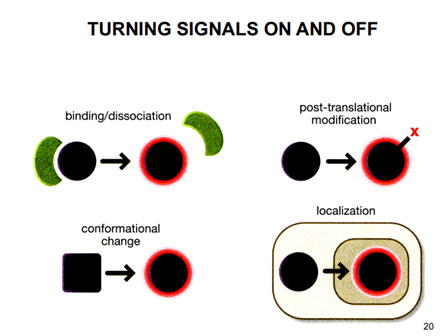
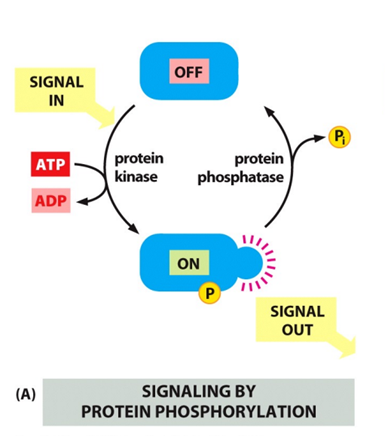
Phosphorylation and dephosphorylation – adding and removal of a phosphate group
Two families:
a) Serine threoninE
b) Tyrosine
- Protein kinase ass on P while protein phosphotase romove the P
- Regulate with ATP hydrolysis
- Affects and controls activity of localisation
- Useful with protein-protein interaction, has the phosphate allows for other protein to dock.
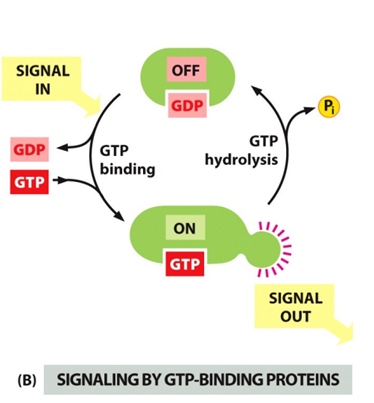
G proteins- binding of GTP or GDP
- Protein is off when GDP is bound.
- Protein is on when GTP is bound
- Dissociation of GDP and binding of GTP.
- Hydrolysis is Accelarted by GEF protins.