2b Proteins and Enzymes
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
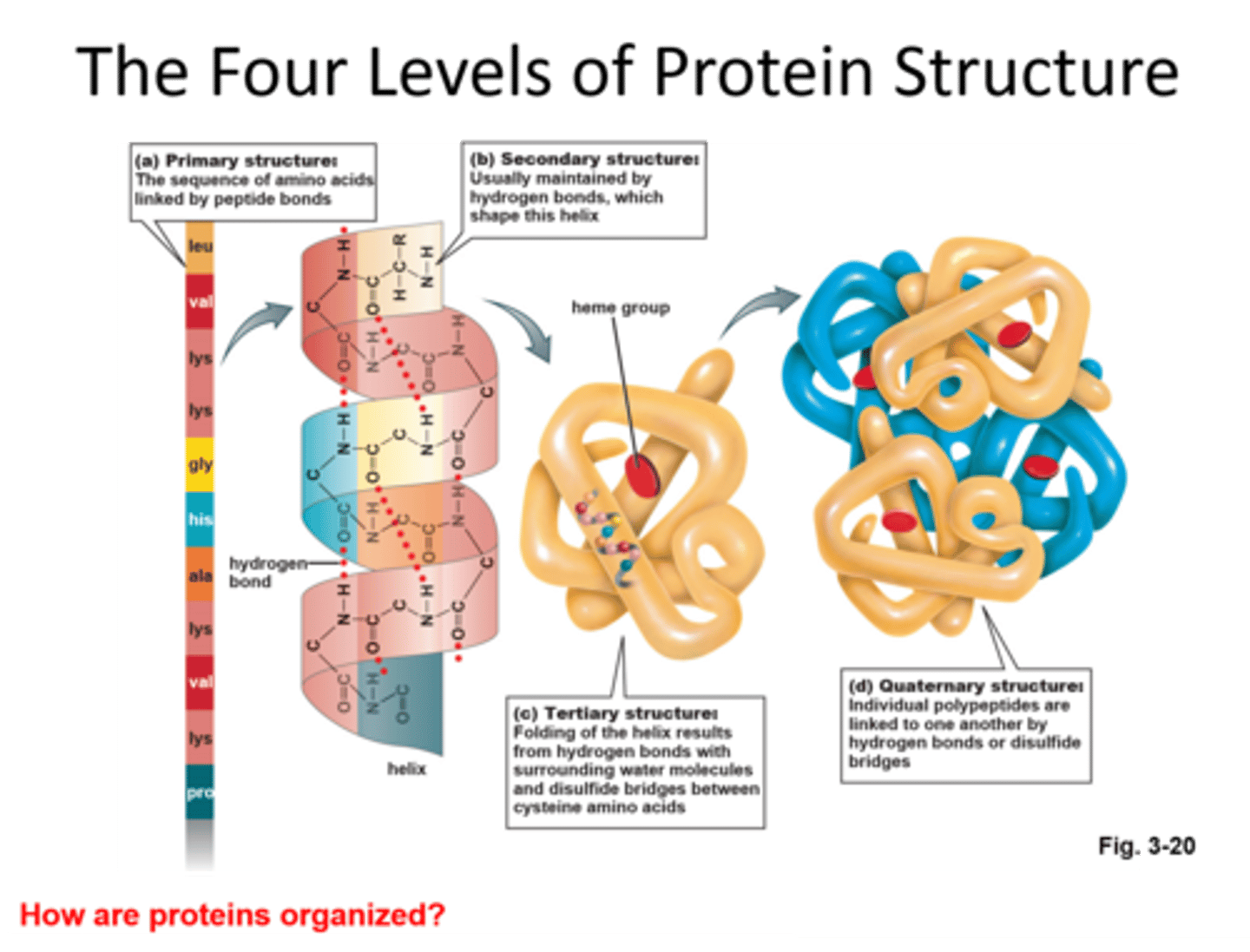
Explain protein structure
· Primary structure - the sequence of amino acids in a polypeptide chain.
· Secondary structure - folding of the polypeptide chain due to the formation of many weak hydrogen bonds (a -helix and the b-pleated sheet).
· Tertiary structure - further folding where the whole chain folds into a 3D specific shape. Stabilised by ionic bonds, hydrogen bonds, and disulphide bonds
· Quaternary structure - made of more than one polypeptide chain, e.g., haemoglobin.
Test for proteins
· Add potassium hydroxide and copper sulphate (or Biuret solution) to a sample of the solution to be tested.
If protein is present a lilac colouration is seen.

Bond formed between two amino acids
Peptide Bond
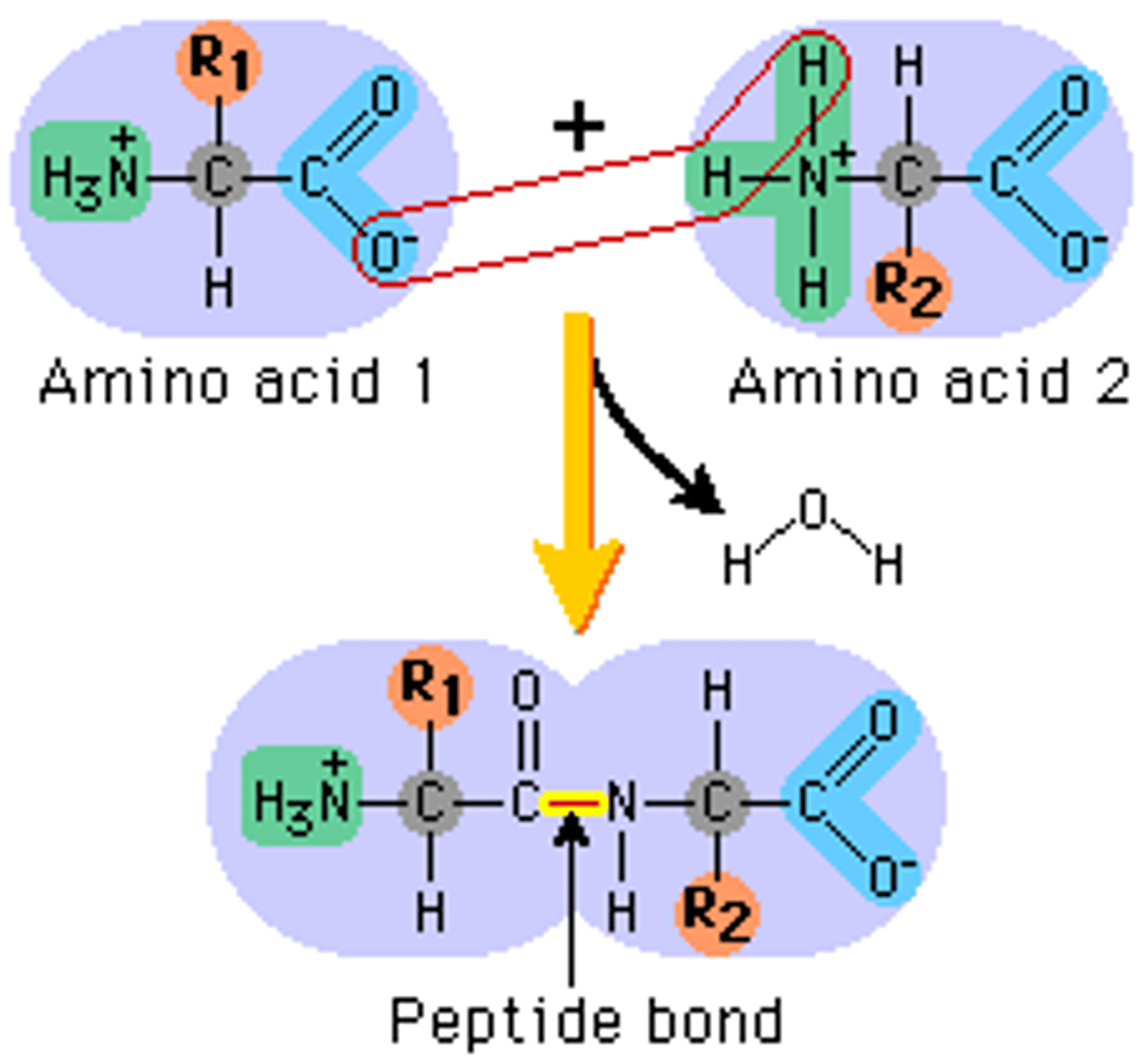
Draw a generalised amino acid

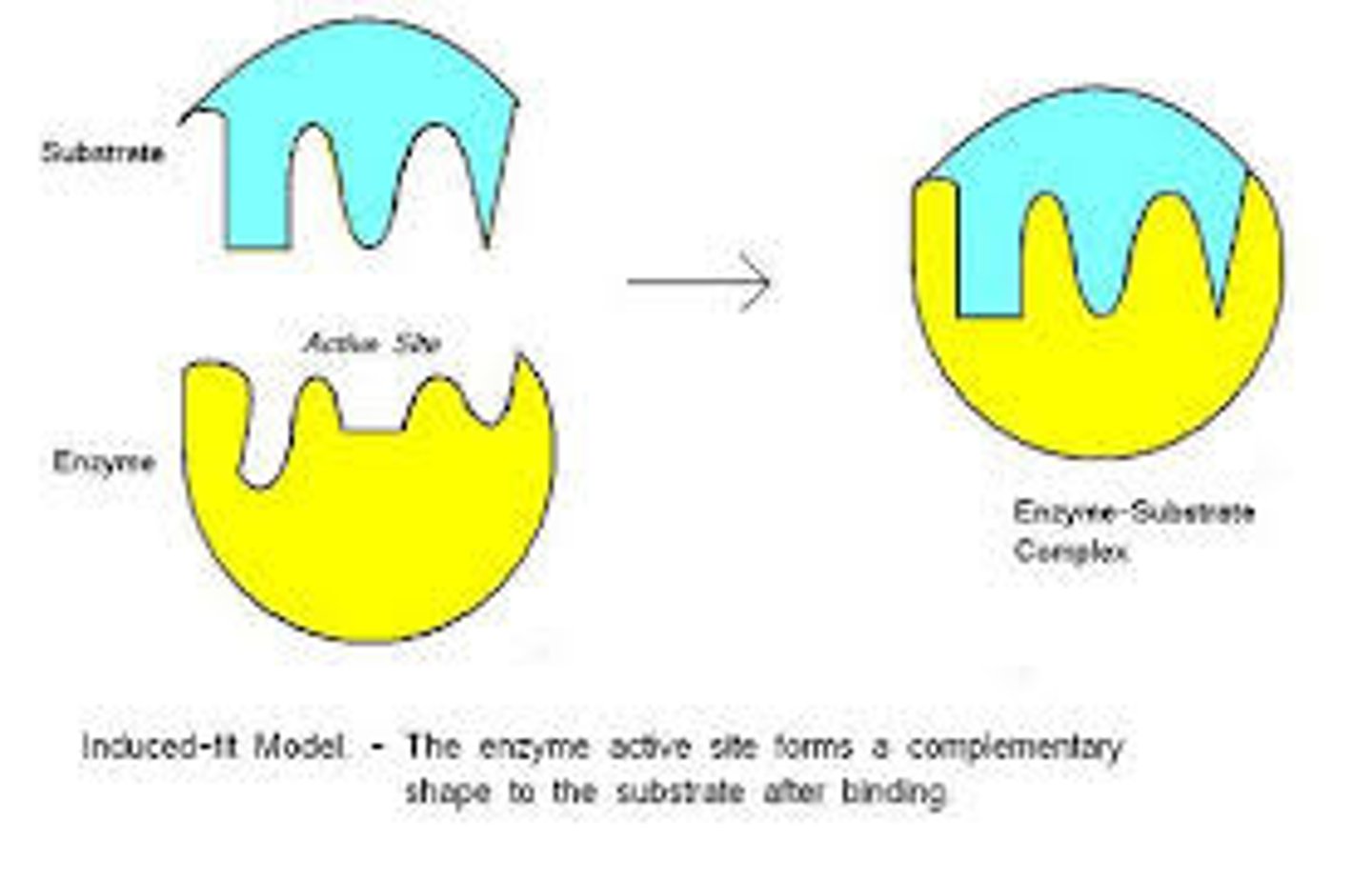
Enzymes work because they have......
· Active site is complementary shape to the substrate
· Substrate binds/fits into active site
· Forms enzyme-substrate complex
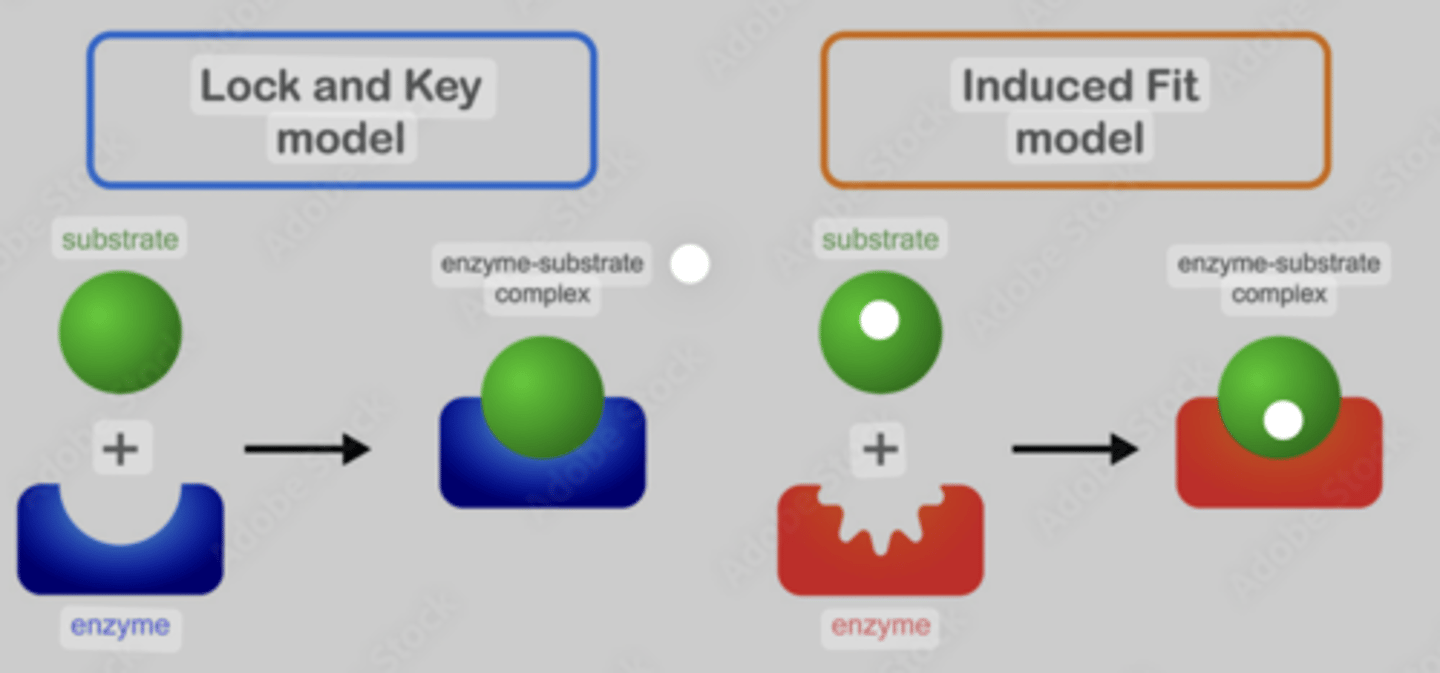
Induced fit model
· A substrate and its active site are not fully complementary to begin with
· When a substrate binds with an enzyme, it induces changes in the enzyme structure.
· The active site changes shape to become complementary.
This puts strain on the substrate and distorts the bonds, thus reducing the activation energy
How does the induced fit model explain enzyme action?
The active site molds around the substrate
This distorts the bonds
Lowering the activation energy
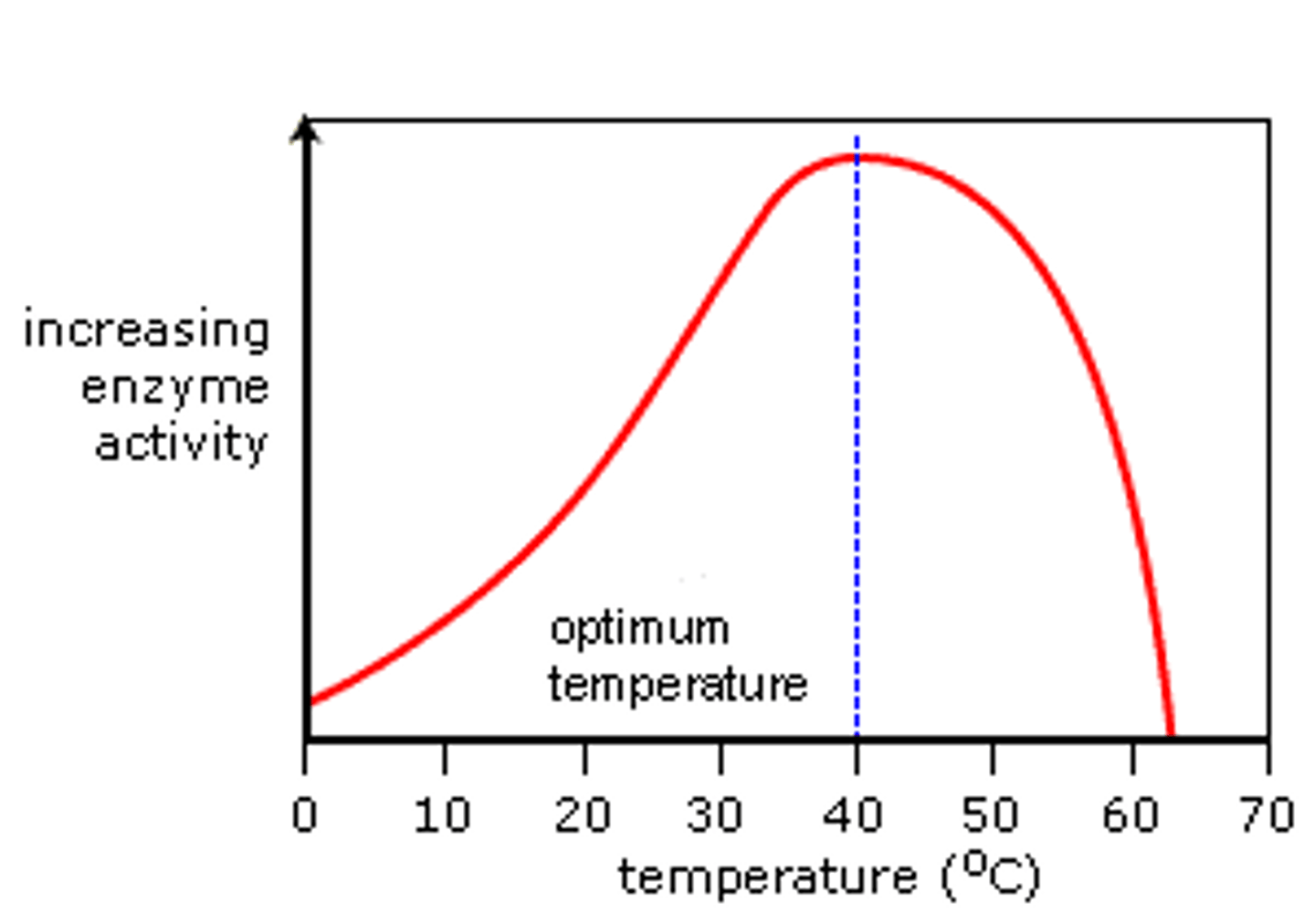
Effect of temperature on enzyme action
· As temperature increases, enzyme and substrate molecules gain more kinetic energy.
· They collide more frequently
· So a greater number of enzyme-substrate complexes are formed
· Leading to an increased rate of reaction.
Effect of pH on enzymes action
· A change in pH causes some of the hydrogen and ionic bonds that maintain the enzyme's tertiary structure to break.
· The shape of the active site changes
· No enzyme -substrate complexes can be formed.
· The enzyme has been denatured

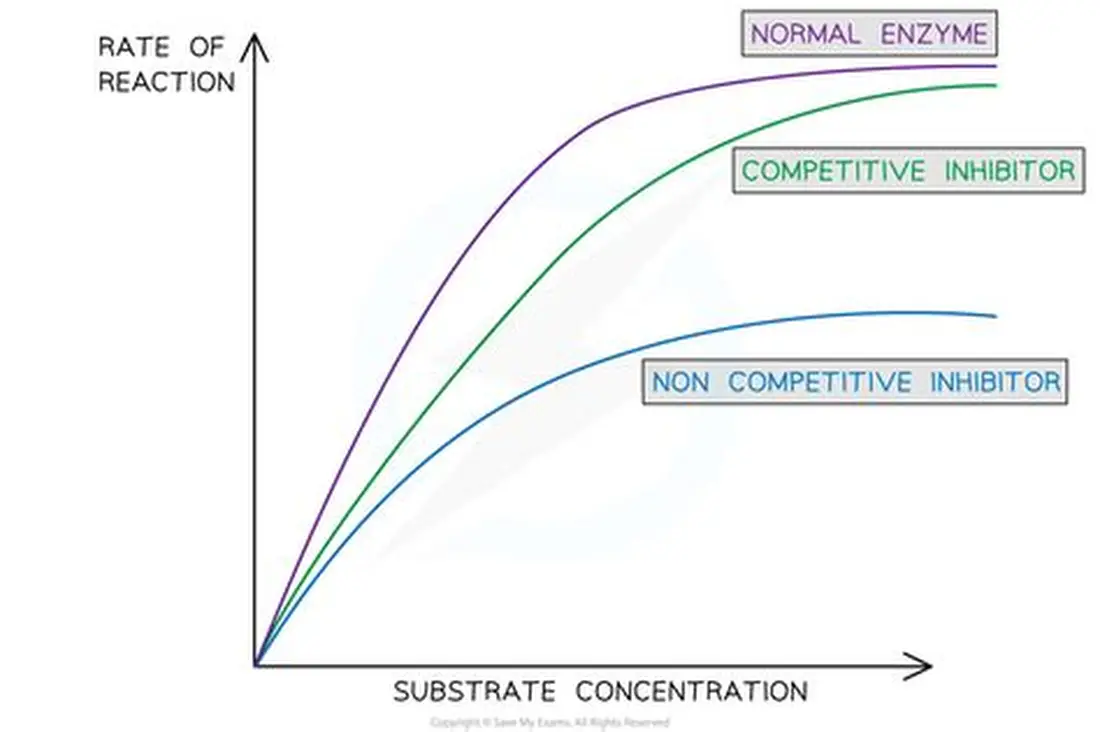
Competitive inhibitors
· Has a similar shape to that of the enzyme's substrate.
· It bind to the active site of the enzyme.
· The active site is occupied and blocked
· Fewer enzyme-substrate complexes are formed and the rate of reaction is reduced.

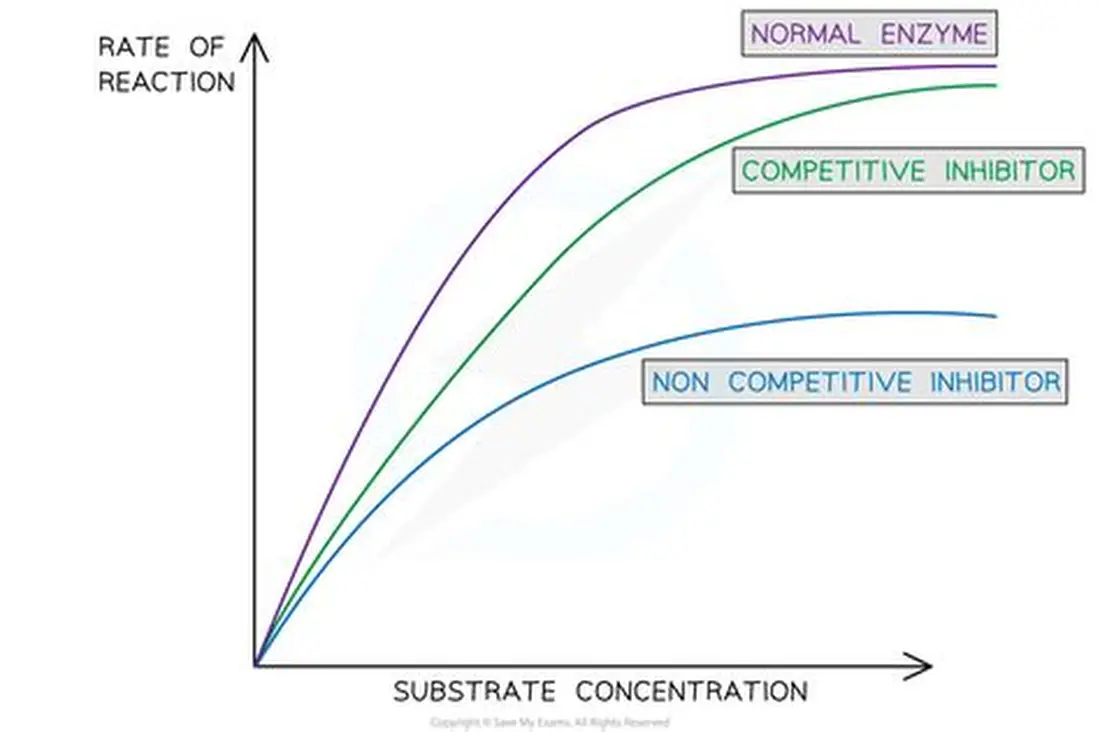
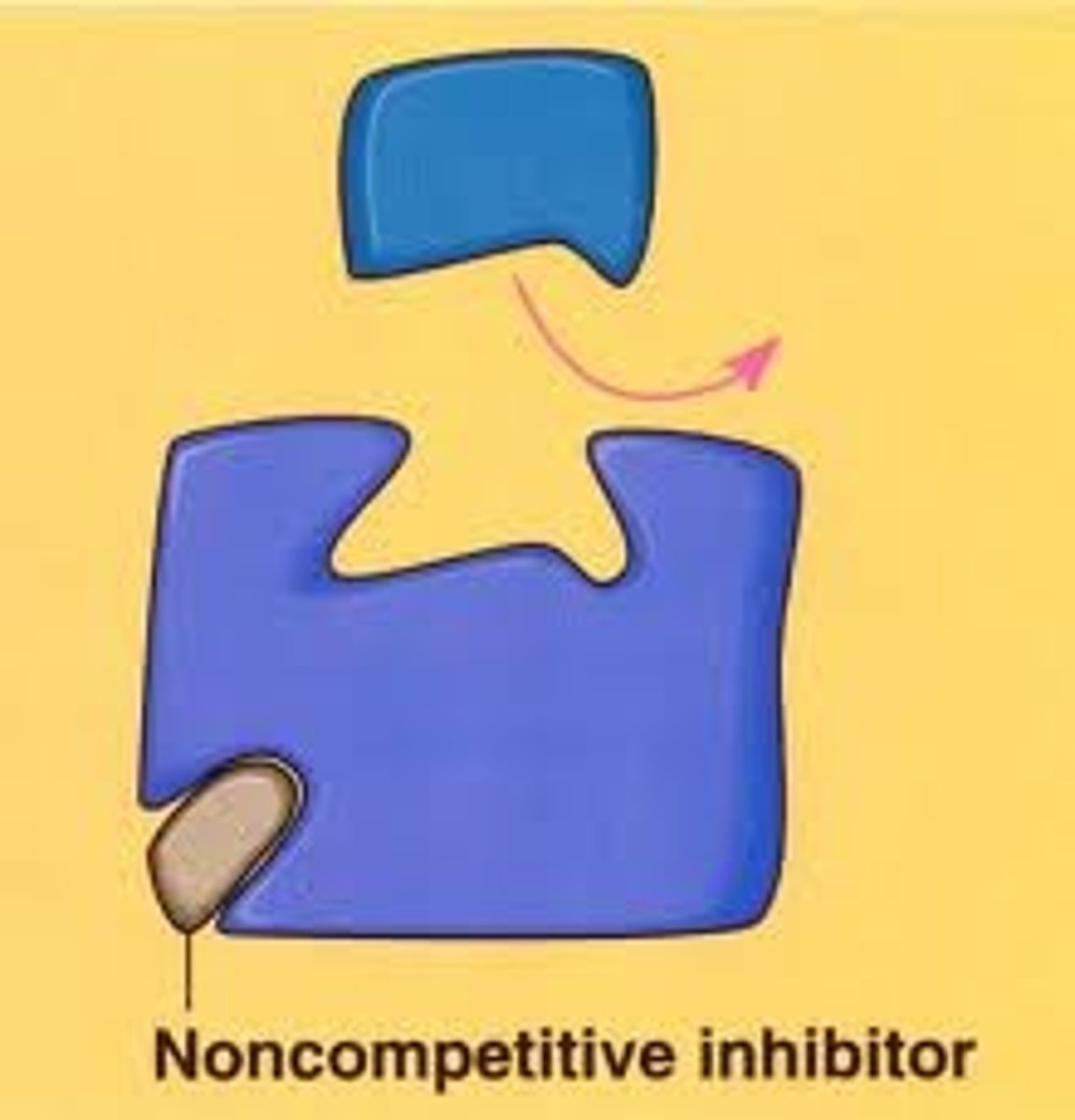
Non-competitive inhibitors
· Non-competitive inhibitors bind to some other region of the enzyme.
· This changes the tertiary structure so changing the shape of the active site
· The substrate cannot bind.
Fewer enzyme-substrate complexesare formed and so reaction rate decreases
What are fibrous and globular proteins?
Fibrous: form long chains running parallel to each other with cross-bridges between the chains- this produces very stable molecules which have a structural role e.g. collagen
Globular: carry out metabolic reactions e.g. haemoglobin or enzymes
Similarities and differences between the induced-fit model of enzyme action and the lock and key model of enzyme action
Similarities: they both form an enzyme-substrate complex, in both the substrate binds to the active site
Differences: initially the tertiary structure of the active site is not complementary to the substrate with the induced fit but is complementary with the lock and key, the structure of the active site changes when the substrate binds but doesn’t change in the lock and key model
What are proteins?
Polymers made up of one or more chain of amino acid monomers joined by condensation reactions, forming polypeptides
Which bonds break first in a change in a protein?
The weakest : hydrogen, ionic, disulphide
Denatures from high temperature or pH
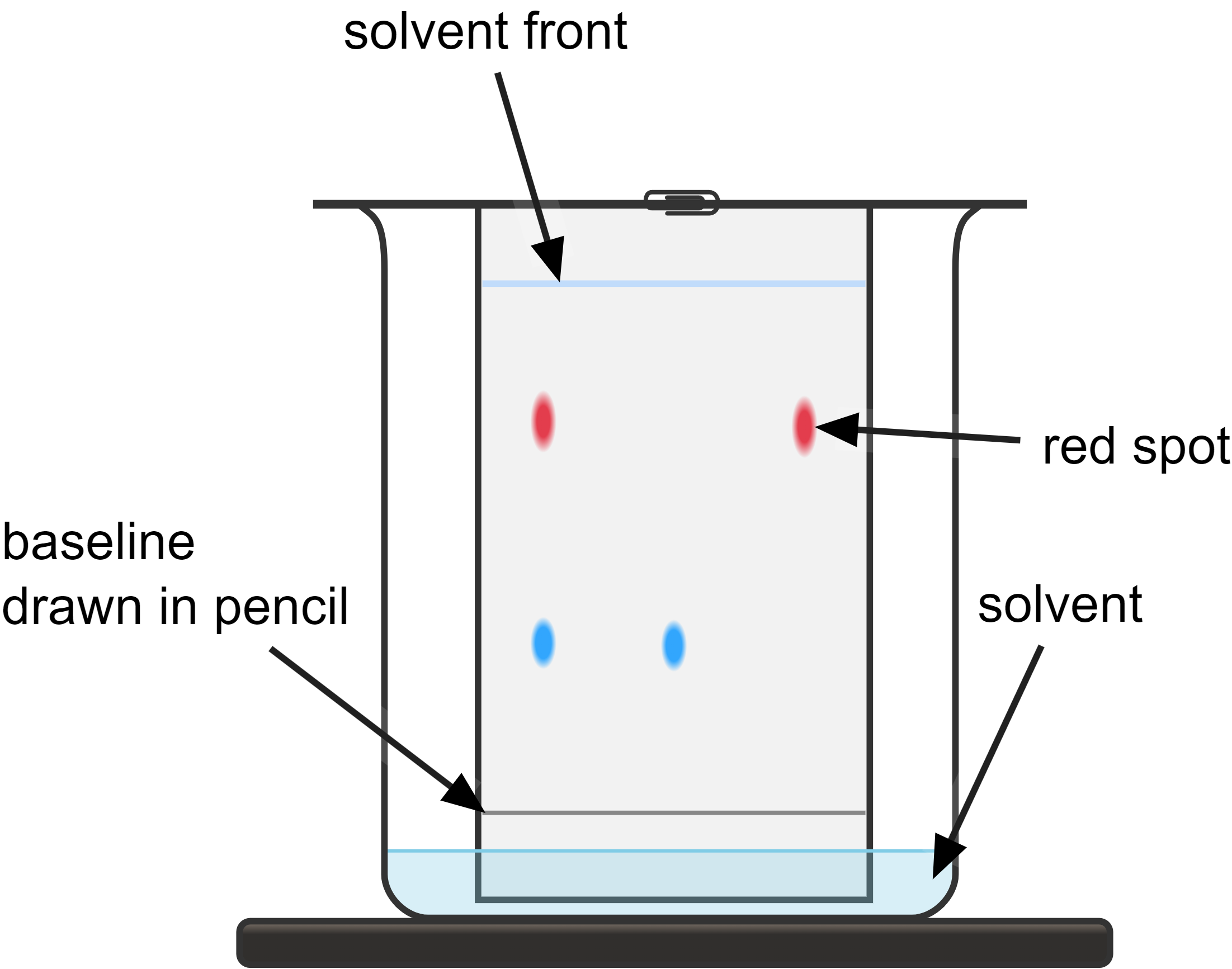
What is chromatography used for?
To separate mixtures of monosaccharides or amino acids
molecules have different sizes and solubilities
smaller and more soluble molecules will move further
Features of enzymes
Biological catalysts that lower the activation energy
All metabolic reactions are catalysed by enzymes
Some are intracellular (inside) and some extracellular (outside the cell)
The active site has a specific tertiary structure
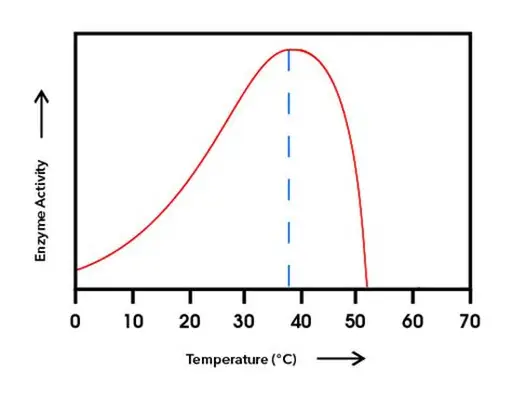
Describe and explain how temperature effects the rate of enzyme controlled reactions, using the graph below
As temperature increases, the rate of reaction initially increases
After the optimum temperature of 37C, the enzyme activity decreases
As temperature increases, enzyme and substrate molecules gain more kinetic energy
They collide more frequently
So a greater number of enzyme substrate molecules are formed
So the rate of reaction initially increases
After the optimum temperature, the increased kinetic energy causes the hydrogen bonds in the enzyme to break
This means that the active site is no longer complementary to the substrate so less enzyme-substrate complexes are formed
The enzyme denatures
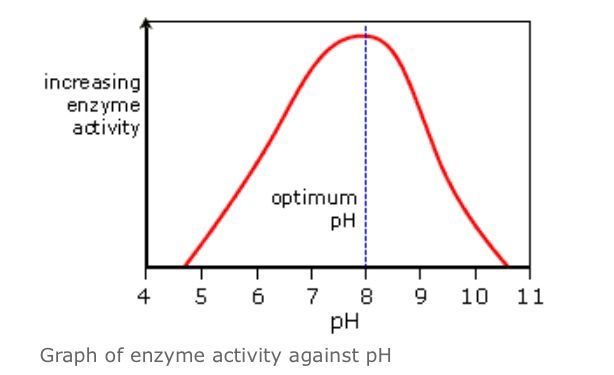
Describe and explain how pH effects the rate of enzyme controlled reactions, using the graph below
Enzyme activity decreases on either side of the optimum pH
The further away from the optimum pH, the greater the decrease in enzyme activity
Low pH: a decrease in the pH away from the optimum increases the number of H+ ions in a solution. The excess H+ ions cause the hydrogen and ionic bonds to break- the enzyme denatures. The active site is no longer complementary to the substrate so less E-S complexes are formed
High pH: an increase in pH away from the optimum increases the number of OH- ions in a solution. The excess OH- ions cause the hydrogen and ionic bonds in the enzyme to break- the enzyme denatures. The active site is no longer complementary to the substrate so less E-S complexes are formed
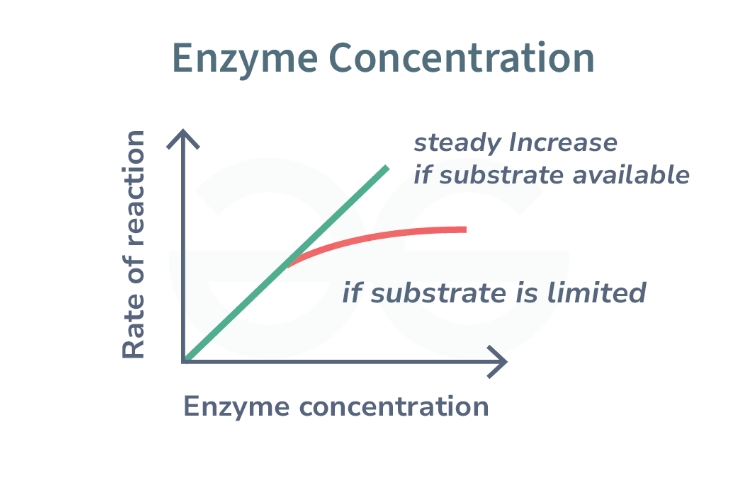
Describe and explain how enzyme concentration effects the rate of enzyme controlled reactions, using the graph below
Initially, enzyme activity increases as enzyme concentration increases
Point reached where further increase in enzyme concentration has no further increase on rate of reaction
With more enzymes in the solution, there are more successful enzyme and substrate collisions (more E-S complexes formed)
If substrate concentration becomes limiting, adding additional enzymes will not result in an increased rate of reaction
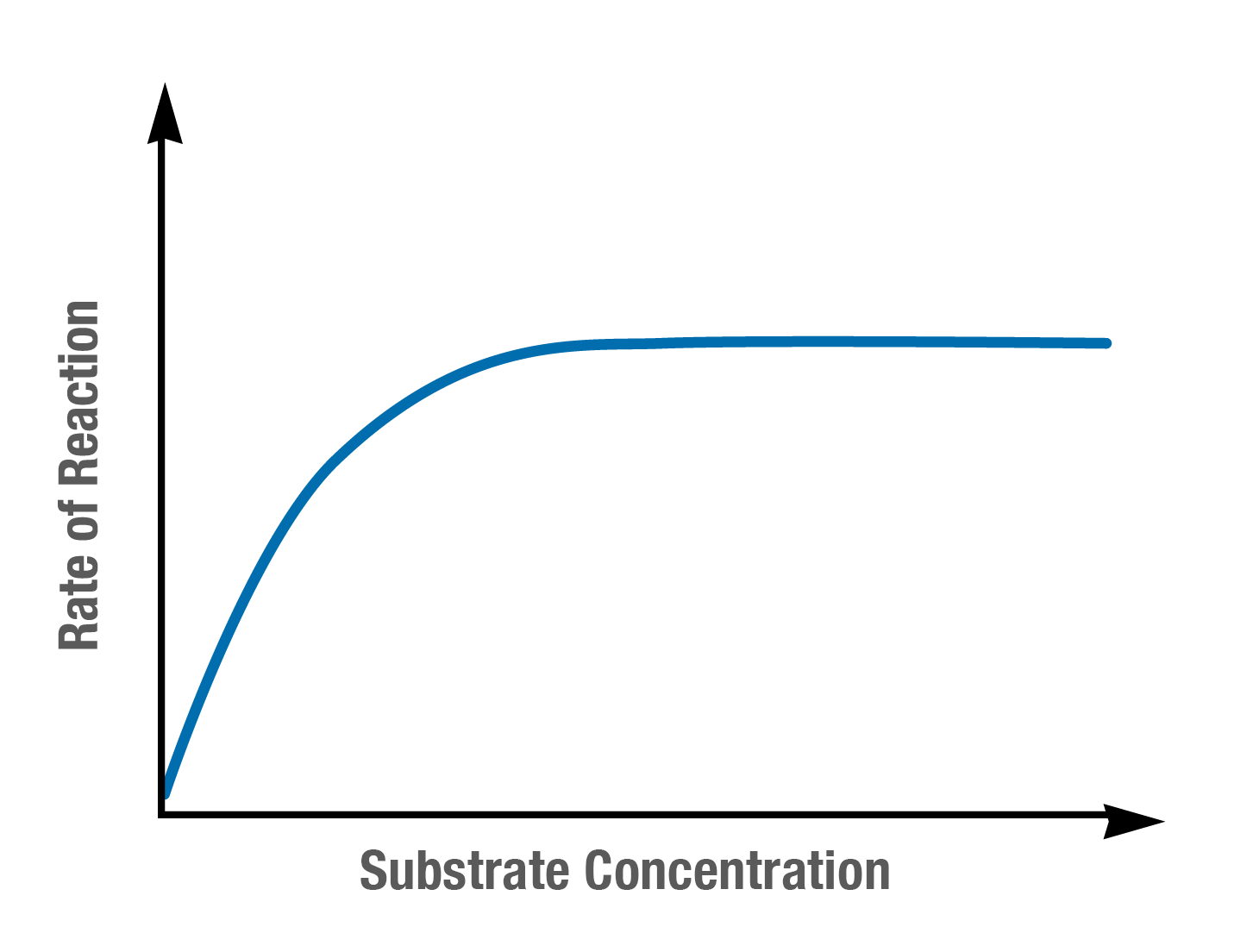
Describe and explain how substrate concentration effects the rate of enzyme controlled reactions, using the graph below
Initially enzyme activity increases as substrate concentration increases
With more substrates in the solution, there are more successful E and S collisions and more E-S complexes are formed
Once the rate of reaction has reached a max, there is no further increase in the rate
All the enzymes active sites are fully occupied, the enzyme concentration is limiting the rate of reaction
RF value
Distance from origin to solute (spot) / distance from origin to solvent front