Bio 2 exam 3
1/128
Earn XP
Description and Tags
Ch 19-22
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
129 Terms
Evolution
population gene pool becomes different over time
causes extinction and origin of species
Evolutionary processes (6)
natural selection
mutation
gene flow
genetic drift
non-random mating
constraints, trade-offs
What did Charles Darwin and Alfred Wallace do?
Evolution by natural selection
species change over time
share a common ancestor and diverge
changes explained by natural selection
Natural selection
increased relative individual fitness of certain individual phenotypes within a population (aka some individuals survive better)
variation in traits
heritable
related to fitness
acts directly on phenotype and indirectly on genotype
Consequence of natural selection
favored phenotypes become dominant in the population and are passed down —> evolution
Allele frequency
proportion of each allele in the population (A vs a)
change in this = evolution
Genotype frequency
proportion of each genotype in the population (AA vs Aa vs aa)
Adaptation
favored traits spread in the population through natural selection
Artificial selection
selective breeding by humans
needs genetic variation for the desired trait
Mutation
genetic variations that provide material for evolution by (natural) selection
is the mutated or WT trait better?
Gene flow
movement of genes in/out of a population
emigration and immigration
Genetic drift
random changes in allele frequencies over generations
mostly impacts small populations
bottleneck and founder effects
harmful allele typically remains
Bottleneck effect
random event where only some survive —> lower genetic diversity
impacts large populations as well
Founder effect
dispersal event causes population to split up into different locations —> lower genetic diversity
Neutral allele/trait
neither beneficial (helpful) nor deleterious (harmful)
Nonrandom mating
individuals prefer to choose some mates over others
if changes genotype frequencies —> no evolution
if changes allele frequencies —> evolution
Homozygous increase
individuals prefer to mate with their own genotypes
Heterozygous increase
individuals prefer to mate with a different genotype
Sexual selection (nonrandom mating)
individuals of one sex prefer to mate with particular individuals in other sexes
only in species with sexual reproduction and 2+ sexes
changes allele frequencies
Intersexual selection
individuals of one sex prefer to mate with individuals of another sex with certain phenotypes
Intrasexual selection
individuals of one sex compete to access mates
only the winner gets to mate
Asexual reproduction
no fertilization
vegetative and parthenogenesis
Vegetative reproduction
clones from non-sexual tissues
Parthenogenesis
genetically variable clones from germ cells
meiosis (typically incomplete) but no fertilization
Sexual reproduction
meiosis in germ cells and fertilization
isogamy, anisogamy, hermaphroditism
Isogamy
similar gametes in both parents
Anisogamy
different gametes: gametic sex
possible sexual dimorphisms by sex determination systems —> sexual selection
Hermaphroditism
sequential and simultaneous (sometimes self-fertilization)
one dominant sex, if dies, other sex changes to dominant
Sexual dimorphism
observable differences in male and female traits of the same species
gametic sex does not always result in clear dimorphism
sex roles are diverse
Constraints
Preexisting physical and chemical limits on evolution (constraint)
lack of genetic variation prevents evolution
Trade-offs
Balance costs and benefits in adaptations
balance must be positive to evolve
Discrete variation
traits affected by one locus are qualitative
Continuous variation
traits affected by more than one locus are quantitative
types of selection apply to quantitative traits
Stabilizing selection
mean/average phenotype is favored by natural selection
after evolution: some variation is lost, mean stays the same
Directional selection
phenotype different from the mean is favored by natural selection
after evolution: mean changes, variation is not lost
Disruptive selection
phenotypes vary in both directions from the mean are favored by natural selection
after evolution: mean stays the same, variation increases
Frequency-dependent selection
maintains genetic variation within populations
2+ phenotypic variations
frequency of each phenotype determines fitness
Advantage of heterozygotes
maintains polymorphic loci
heterozygotes outperform homozygotes under variable environmental conditions
Geographic variation
maintains genetic variation in geographically distinct populations
different local selection pressures for each population
Phylogeny
the evolutationary history of relationships among organisms
Phylogenetic tree
represents historical relationships among lineages
Binomial nomenclature
Used to name organisms, 2 part names in Latin (genus + species)
proposed by Carolus Linnaeus
Naming rules
capitalized genus + lowercased species + who made the name
Taxon
any group of organisms at the same taxonomical level
Clade
a taxon including all evolutionary descendants (node to tips)
Monophyletic group
common ancestor + all descendants
AKA a clade
Polyphyletic group
does not include direct common ancestor
come from different clades
Paraphyletic group
common ancestor + some descendants
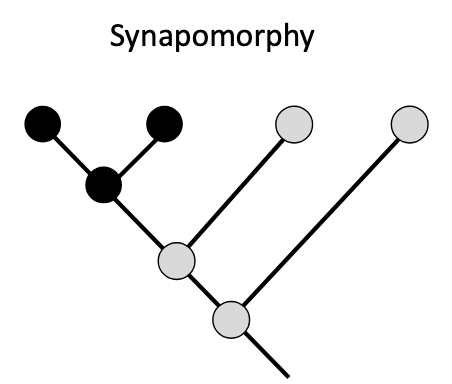
Synapomorphies
shared derived traits
ex. gizzard in both crocodiles and pigeons
Homologous traits
share similar evolutionary origins
ex. similar bone shapes/structures in bat vs. bird wings
Analogous traits
evolve through convergent evolution
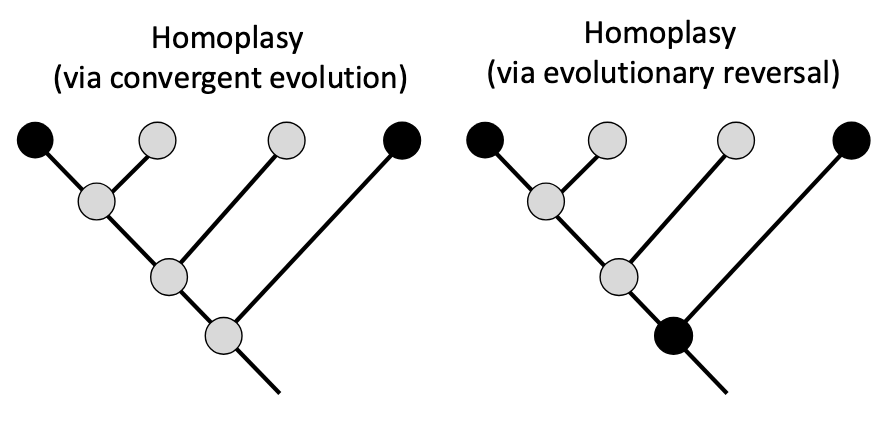
Convergent evolution
evolving independently for the same function
Evolutionary reversal
trait goes back to its original function
ex. fins in dolphins > hands in humans
Homoplasies
similar traits that come from convergent evolution or evolutionary reversals
Molecular clocks
use mutation rates to label dates for evolutionary events
rate of change/slope of the graph line
Phylogenetic trees can be built based on ?
synapomorphies
Principle of parsimony
the best phylogenetic tree requires the fewest evolutionary changes (lowest # homoplasies)
Consensus (phylogenetic) tree
computed because results are different depending on the traits used
Mathematical models
required to build phylogenetic trees from extensive molecular data (nucleotide sequences)
more flexible than parsimony
maximum likelihood
Comparing gene and protein sequences to identify evolutionary changes
compare aligned homologous sequences
mutations in DNA accumulate at different rates over time
(rate = molecular clock)
regulatory sequences accumulate less mutations than protein-coding regions
exons accumulate more mutations than introns
Observed vs. actual number of changes between aligned sequences
Observed # underestimates actual # from common ancestral sequence
multiple substitutions would be undetectable
calculate evolutionary divergence
Evolutionary divergence
calculate by considering possible rates of change in mathematical model
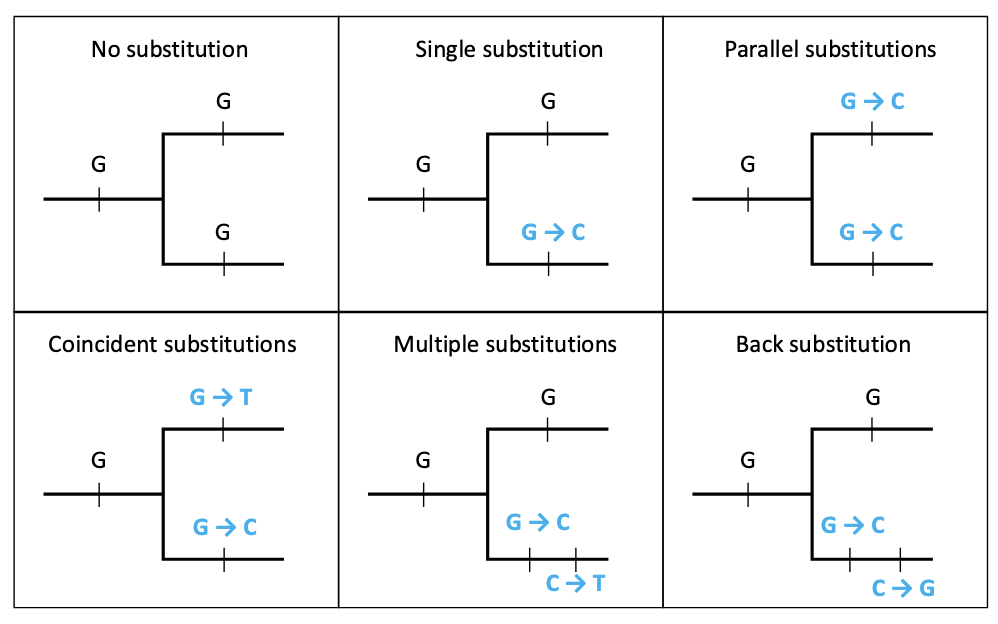
Different types of substitution
None, single, parallel (2+ same change), coincident (2+ different change), multiple (2+ different change in same sequence), back (evolutionary reversal)
Synonymous substitutions (silent)
neutral to natural selection, accumulate over time
*mutation rate is same as non-synonymous
Non-synonymous substitutions
under natural selection
mostly deleterious/removed
some almost neutral
some beneficial/favored
Selection in genes among different populations/species
detect by comparing rate of synonymous and non-synonymous substitutions
Rate of synonymous/non-synonymous substitutions
~1 = neutral
syn < non-syn = positive selection
syn > non-syn = purifying selection
Genome sizes
differ by orders of magnitude
prokaryotes < eukaryotes
larger =/= greater complexity
large genomes have more non-coding DNA
selection against non-coding is stronger in large populations
Sexual reproduction
results in recombination of genomes by crossing over during meiosis and combination of gametes
Sexual reproduction disadvantages
recombination breaks up adaptive gene combinations
sex reduces # genes each individual passes
Sexual reproduction advantages
repair damaged DNA
purging deleterious mutations
new combinations of alleles
Lateral gene transfer
horizontal movement of genes between lineages that results in gain of new functions
increased genetic diversity, new functions in new DNA
bacterial transformation, transposons, hybridization of species
Reticulations
events of horizontal gene transfer, includes inheritance proportion #
Eukaryote gene families
result of gene duplication
evolutionary tinkering
concerted evolution
Evolutionary tinkering
copies diversify in form and function; 1 original copy and other mutated copies
mutations can be beneficial/favored or not/pseudogenes
Concerted evolution
copies do not diversify in form and function
unequal crossing over
biased gene conversion
Unequal crossing over
misalignment of repeats > crossing over > increase in copies
can quickly spread or eliminate copies with new substitutions
Biased gene conversion
one gene copy damaged > repair with template from favored sequence > extra favored copy is added to original chromosome
Cascade of TFs
establishes body segmentation in animals
maternal effects genes
segmentation genes
hox genes
Maternal effects genes
mRNA and proteins, sets up anterior-posterior axes of the body
Segmentation genes
determine boundaries and polarity of each segment
gap genes: organize broad areas on anterior-posterior axis
pair rule genes: divide embryo into units of 2 segments
segment polarity genes: determine boundaries and anterior-posterior organization
Hox genes
determine role of each segment; what organ develops at each location
encode TFs to determine role
homeobox
Homeobox
conserved sequence in hox genes that codes for a homeodomain (in TF)
binds to DNA to regulate gene expression
Homeotic mutations
in hox genes, can replace organs
Genetic toolkit of hox genes
we all share a conserved developmental toolkit
similar chromosomal arrangement
ordered as they are expressed
Developmental modules (ex. body parts)
can evolve independently within a species; changes in forms and functions
genetic switches
Genetic switches
control how the genetic toolkit is used (on/off genes)
Modularity
allows independent evolution of developmental processes
heterometry
heterochrony
heterotopy
mutations of developmental genes
Heterometry
amount of gene expression
Heterochrony
timing of gene expression
Heterotopy
location of gene expression
Sexually reproducing species
group of actually/potentially interbreeding natural populations that is reproductively isolated from other groups
Speciation
divergence of lineages and emergence of reproductive isolation between lineages
Dobzhansky-Muller model
explains evolution of reproductive isolation
ancestral lineage can separate into 2 if there is a barrier to gene flow (ex. geographic isolation)
descendant lineages now evolve independently
can experience allele fixation over time
barrier can disappear and allow lineages to interbreed, but they are now incompatible and can cause low fitness/death
Allele fixation
loss of polymorphism result in just 1 allele in a locus (ex. A is fixed and a disappears, so only AA)
Reproductive isolation
increases with genetic divergence, is not all or nothing
Reproductive compatibility
decreases with geographic isolation and genetic divergence
greater difference with greater mutation rate
Speciation by centric fusion
normal chromosome pairing is not possible in hybrids after centric fusion
D-M model is applied to chromosomes instead of loci
takes only a few generations in comparison to reproductive isolation, which takes millions of years
Centric fusion
chromosomal rearrangement
Allopatric speciation
mutation, genetic drift, local adaptation due to physical barriers that separate a population OR founder effects when a group crosses an existing barrier
most common mode of speciation (but slow)
lineages split, reproductive isolation (D-M)
pairs of extant closely-related sister species