acetylation and disease + ubiquitnation
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
what AA can a acetyl group be added to
lycine K
asparagine R
glutamine Q
•Acetylated proteins most abundant in mitochondria
what are the 2 types of acetylation
irreversible - N terminal
reversible - lysine
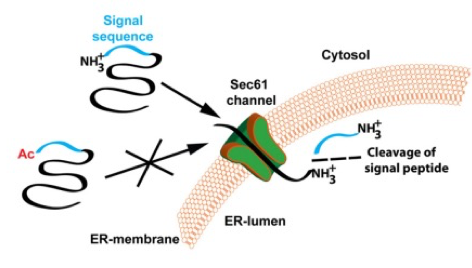
Irreversible
N terminal of methionine is cleaved by N terminal acetyltransferase (NAT) and replaced by an acetyl group from acetyl co enzyme A
subcellularly
reverisble
activates/deactivates proteins
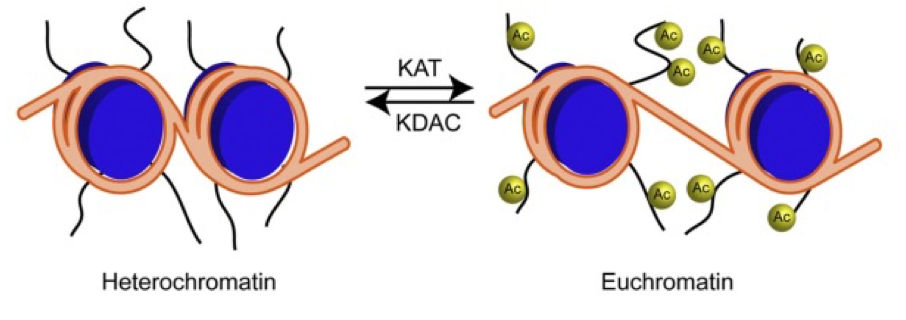
adition of acetyl to lys by histone/lysine acetyltransferase (HAT/KAT)
•acetylation of histones promotes transcription by reducing chromosomal condensation
•
•Removal – histone/lysine deacetylases(HDAC/KDAC)
what does protein acetylation regulate
protein stability - can protect proteins from degradation or promote degradation via the ubiquitin-proteasome system.
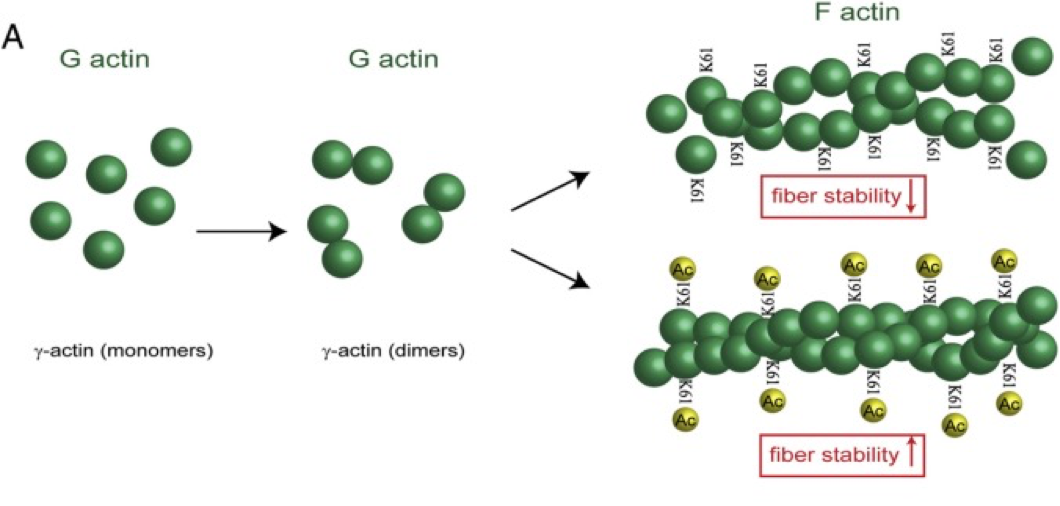
Cytoskeleton remodelling
cell cycle
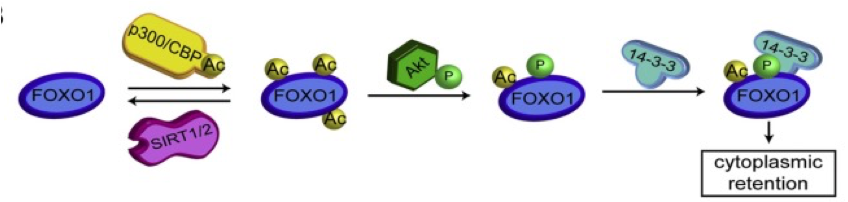
regulated processes
•RNA splicing
•Nuclear transport
•Mitochondrial biology
•Chromatin remodelling
•Transcription
- Protein-protein interactions
- Protein-DNA interactions
what is ubiquitination
ubiquitin, a small regulatory protein, is covalently attached to a target protein.
is a 76 AA regulkatory protein
is added to lys residues of protein

•How cells interprets ubiquitination signal depends on 2
•Number of ubiquitin molecules added - Mono or poly-ubiquitinated
•How ubiquitin molecules linked together - Linkage through Lys48 or Lys63
types of ubiquitination

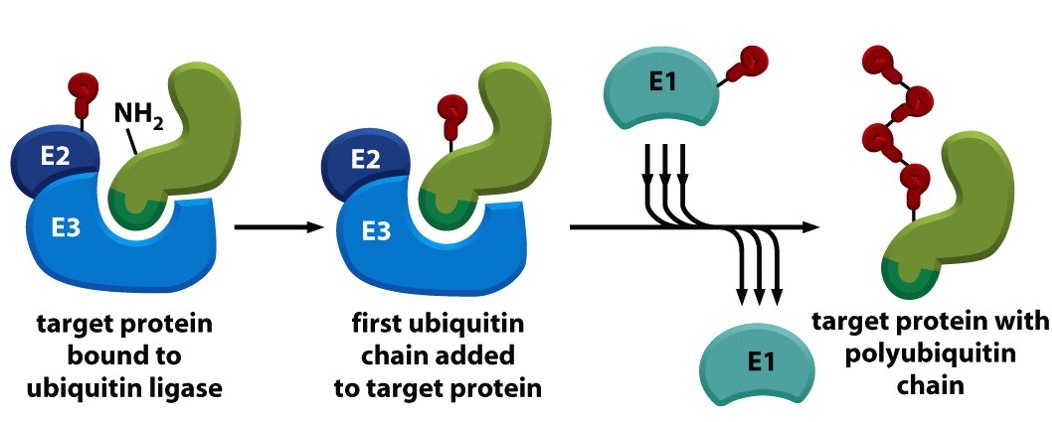
Mechanism of Ubiquitination
Activation (E1 - Ubiquitin-activating enzyme)
Ubiquitin is activated in an ATP-dependent manner and forms a thioester bond with E1.
Conjugation (E2 - Ubiquitin-conjugating enzyme)
The activated ubiquitin is transferred to the E2 enzyme.
Ligation (E3 - Ubiquitin ligase)
E3 transfers ubiquitin from E2 to a specific lysine residue on the target protein, determining substrate specificity.
what is proteolysis
a highly regulated process in which inactive precursor proteins (zymogens or proenzymes) undergo proteolytic cleavageto become active enzymes.
what is proteolytic cleavage
the enzymatic process by which specific peptide bonds in a protein are hydrolyzed, leading to activation, maturation, or degradation of the protein. T
proteolytic cleavage PTM’s
•Activation
•Inactivation
•Completely altered protein function
•Removal of signal peptide
why are proteases synthesized as inactive zymogens
to prevent premature activity
what are caspases
•Cysteine-dependent aspartate-directed proteases
cleave proteins at aspartic acid
what is caspases inactive zymogens called
procaspases
•Need proteolytic cleavage to get activated
what do initiator caspases activate
effector caspases
executes apoptosis
what proteins do caspases target
actin, DNA repair proteins, DNase inhibitor
what happens to the cell content after apoptosis
(proteins, DNA) fragmented and packed into apoptotic bodies
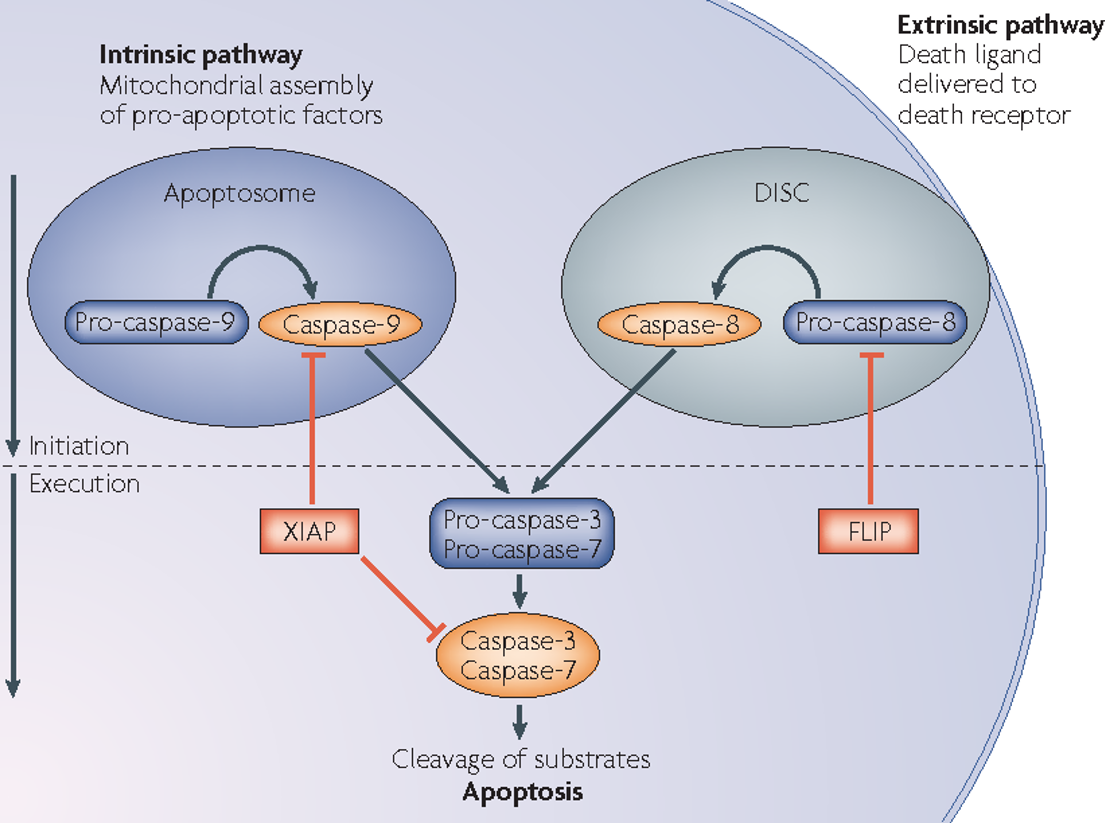
extrinsic pathway for apoptosis
Triggered by death ligands binding to death receptors
Caspase-8 is activated and subsequently activates executioner caspases (Caspase-3, -6, -7).
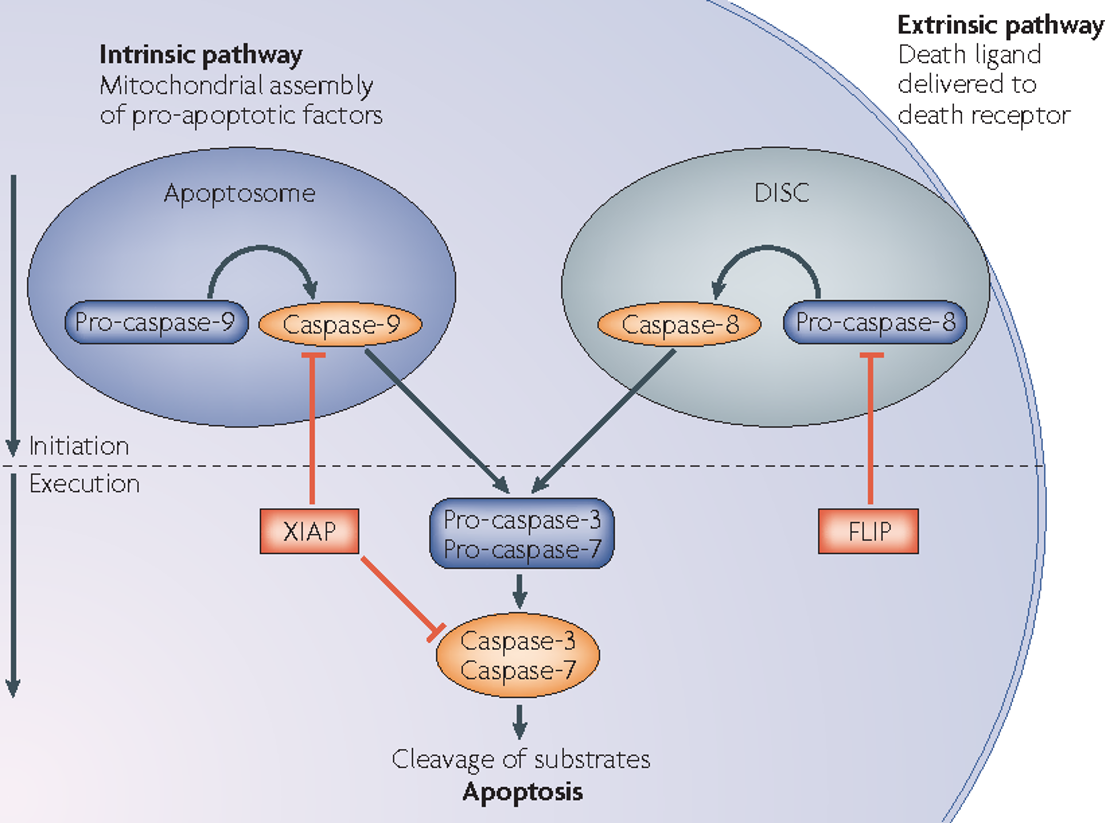
intrinsic pathway for apoptosis
Triggered by DNA damage, oxidative stress, or growth factor deprivation.
Caspase-9 is activated, leading to executioner caspase activation
what gene encodes tumour protein p53
TP53
most prominent tumour suppressor gene
what is tumour protein p53 and what is it regulated by
a transcription factor that prevents mutated DNA being inherited
•Regulated by phosphorylation, methylation, acetylation, sumoylation and ubiquitination
what causes p53 to be activated
dna damage
oncogene activation
hypoxia
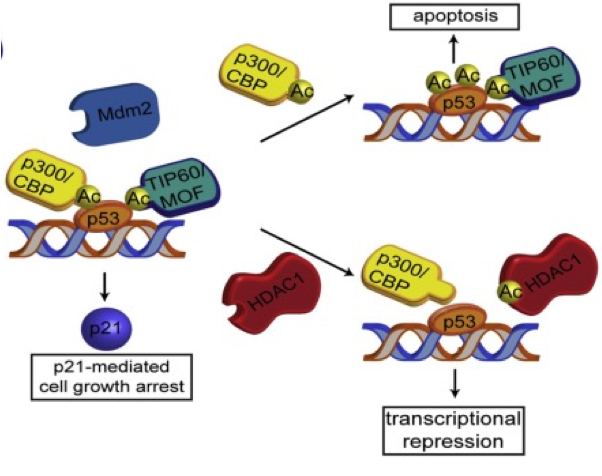
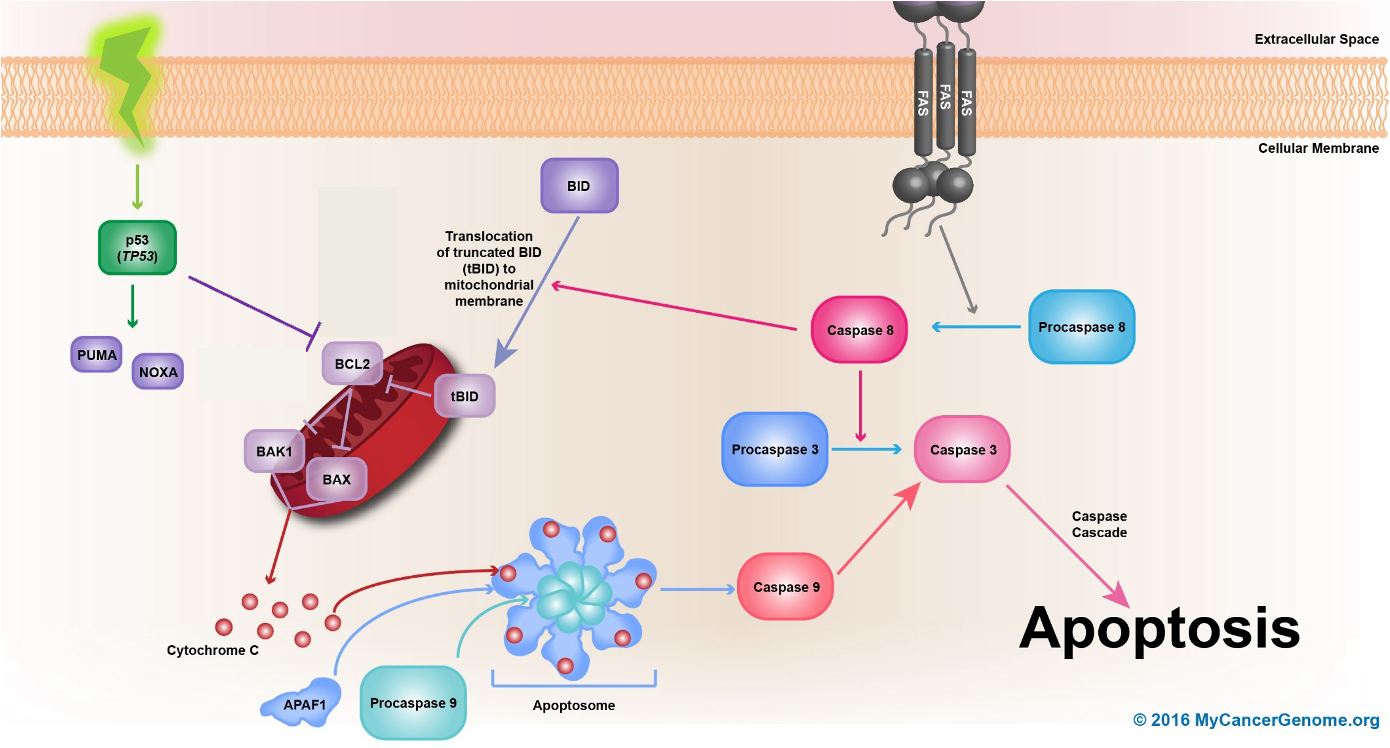
•Upon detection of DNA damage p53 gets activated initating
• transcription of p21 gene leading to cell cycle arrest (G1/S)
•Activates DNA repair genes
•If DNA cannot be repaired p53 induces apoptosis by
- activating pro-apoptotic genes PUMA, NOXA, BAX, FAS
- repressing anti-apoptotic gene Bcl-2
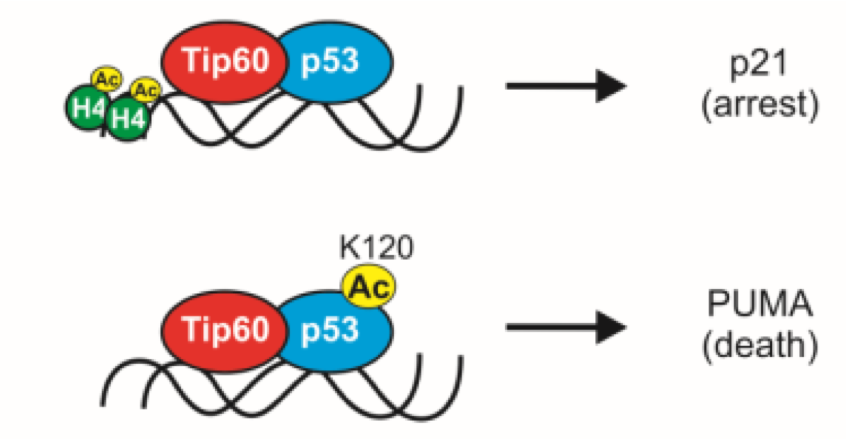
what happens when the dna is damaged
•p21 transcription independent on K120 status à cell cycle arrest
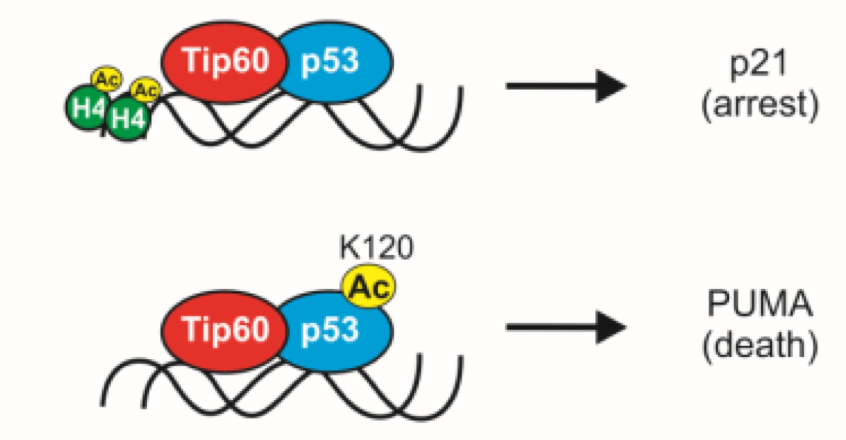
what happens when there is excessive DNA damage
• acetylation of K120 à p53 accumulates at promoters of apoptosis-inducing genes
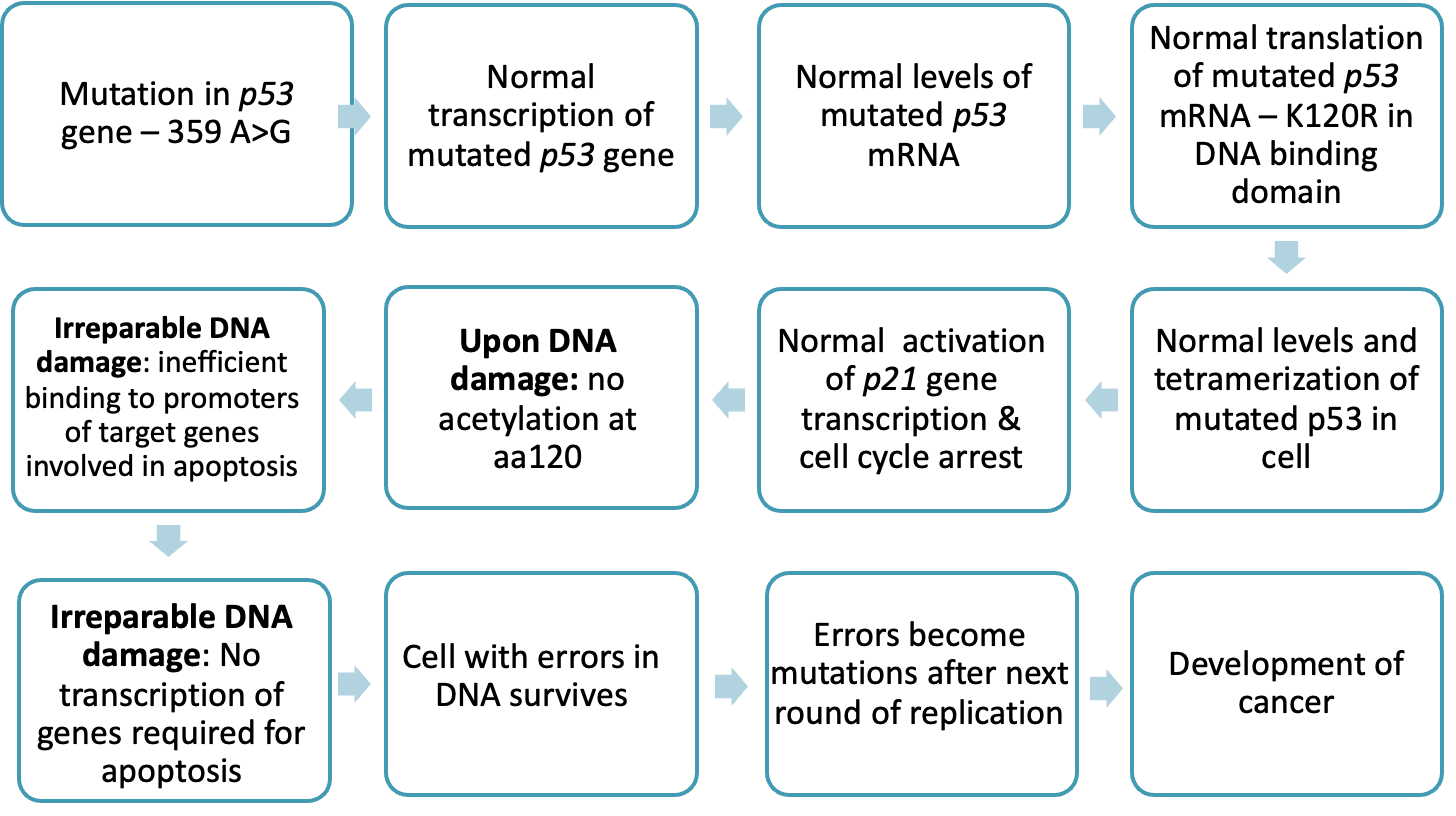
what TP53 mutations are there
•Missense mutation – 359 A>G
•Lys substituted by Arg at aa position 120 (K120R)
what does the K120R mutation cause
•Acetylation of aa 120 impossible
•Ineffective binding to promoters of pro-apoptotic genes
•TP53-mediated apoptosis does not occur
•Cells with errors in DNA survive and proliferate
•Development of cancer
acetylation and disease summary