Structure of Nucleic Acid and Synthesis of DNA
1/52
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
53 Terms
Describe the organization of the genome for prokaryotes, virus, plasmids,
prokaryotes: no special subcellular structure; located in the nucleoid
Viruses: has DNA or RNA, no system for replication, transcription or translation
Plasmids: small circular DNA molecules; can replicate autonomously
Describe the organization of genome for Eukaryotes
has a nuclear envelope
DNA associated with Histones = chromatin
diffuse - euchromatin (active)
condense - heterochromatin (silent)
what are chromatids?
when DNA is replicated and two identical sisters are formed (chromatids)
describe the structure of DNA
nucleosides joined with inorganic phosphate
phosphodiester bond between 3’ carbon of one sugar to the 5’ carbon of the next sugar
describe the A form and Z form of DNA
A: wider right hand spiral
occurs in dehydrated samples of DNA, in DNA-RNA hybrids, and in enzyme-DNA complexes
Z:
linked to initiation of transcription and extensive methylation
z-binding protein may be invovled in regulation of transcription
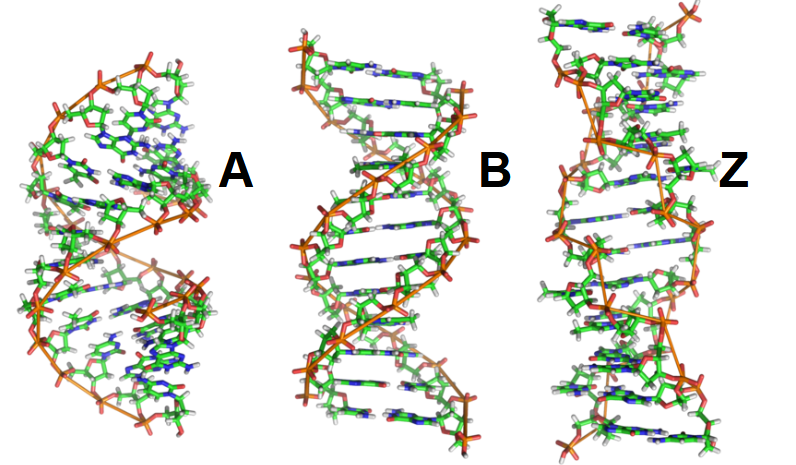
what is the most common form of DNA and describe it
B-form
axis of symmertry
chains are antiparallel
has major and minor grove
regulatory proteins, anticancer drugs and antibiotics bind here
how can strands of DNA be seperated?
by akali
breaks down phosphodiester bonds in RNA
Heat
converts dsDNA to ssDNA
this is called denaturation or melting
Tm = temp where 50% of dsDNA is seperated
depends on GC content, more GC = more stable
what is renaturation/hybridization? How is this used in different molecular biology techniques?
if temperature is slowly decreased, the ssDNA combines to dsDNA
Used in:
Flourescent in situ hybridization (FISH): detect specific sequences
Southern blot (DNA) and northern blot (RNA): detects sequences after seperation in gel
PCR
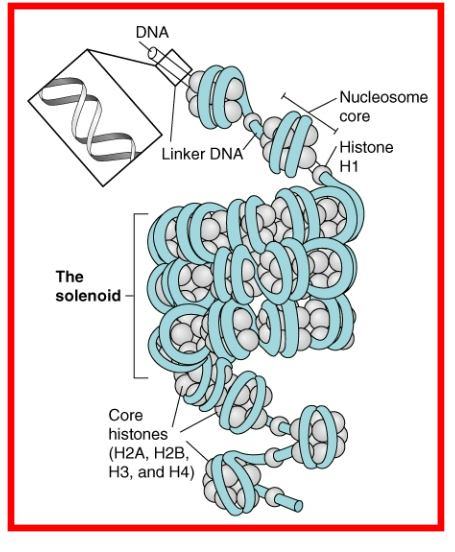
in prokaryotes, how is DNA packaged?
DNA is supercoiled and attached to RNA-protein core forming nucleoid
in eukaryotes, how is DNA packaged/organized?
via binding of histones (H1, H2A, H2B, H3, H4)
nucleosome core formed via 8 histones molecules;
nucleosome combine to produce tubular structure called Solenoid (30 nm fiber)
these 30 nm fiber (aka chromatin fiber) is then organized into loops and anchored via nuclear scaffold proteins
linker dna = histone 1
Describe the location of these chromosomes in humans
it is found that each individual chromosomes occupy different locations in the cell nucleus,
what are the five posttranslation modification on Histones
acetylation
methylation
ubiquination
sumoylation
phosphorylation
what does acetylation do to nucleosomes?
acetylation of lysinee → decreased net positive charge→ decreases histone-DNA interaction → open chromatin;
these are done by HATs (histone acetyltransferase) which coactivator proteins have. These are activated by positive transcription factors
describe histone methylation
histone methyltransferases methylates H3 on lys4, 9, 27 and 79.
methylation have different biological effects
what are genetic locus and alleles?
genetic locus: position of a gene on a chromosome
alleles: alternative versions of a gene found on different homologous chromosomes
what is karotyping?
technique used to determin the diploid number of chromosomes
what is turner syndrome?
single x chromosome in females
universally infertile
kidney/cardiovascular problems
lack of estrogen = osteoporosis
What is Klinefelter syndrome?
most common male disease: XXY
infertile, gynecomastia (enlargment of breast tissue in males)
germ cell/ male breast cell cancer
What is edwards disease?
trisomy of chromosome 18
Kidney malformations and structural heart defects.
Intestines protruding outside the body and developmental delays
What is down syndrome?
trisomy of chromosome 21
What is patar syndrome?
trisommy of chromosome 13
Can results in microcephaly, heart and kidney defects, and abnormal
gemitalia
list two RNA molecules that can catalyze reactions
RNAase P: cleaves off extra RNA sequences at end of tRNA
peptidyl transferase: activity of ribosome
what is the difference between prokaryotic and eukaryotic ribosomes
Prokaryotes: contain only three types of rRNA and less proteins.
Cytoplasmic eukaryotic ribosomes contain four types of rRNA and more proteins.
describe snRNA
A component of small nuclear ribonucleoproteins (snRNPs or
“snurps”).Involved in maturation of RNAs precursors - splicing
describe miRNA (microRNA)
Small non-coding RNAs ~20-24 nucleotides
Derived from endogenous genes
Repress protein biosynthesis, usually by preventing the ribosome
binding to mRNA or promotes mRNA degradation.
describe long noncoding RNA (LncRNA)
200 bp long
Epigenetic modifications of DNA
Example: Xist RNA (X-inactivation specific transcript) inactivates one of the two X chromosomes in vertebrates.
describe Large intergenic non-coding RNAs (lincRNAs)
>200 bp long
Example: scaffold DNA-chromatin complexes
lincRNA HOTAIR is overexpressed in breast tumors
how can DNA synthesis be inhibited and give examples
by incorporation of certain nucleotide analogs.
Replacement of hydroxyl group by azido group Zidovudine (AZT)
reverse transcriptase has higher affinity for AZT then normal human DNA polymerase
Removal of hydroxyl group from deoxyribose Didanosine (ddI)
Conversion of deoxyribose to another sugar, arabinose.
what does Azacitidine (Vidara) do?
chemical analogue of cytidine
Used to treat myelodysplastic syndrome.
Can be incorporated into RNA and DNA.
In RNA, it interferes with polyribosome assembly and tRNA recognition
In DNA, it covalently binds to DNA methyltransferases, which interferes with DNA synthesis and triggers DNA repair and degradation of methyltransferase.
Recently showed promising results in the treatment of gliomas
What does 5’- flourauracil do? (5-FU)
inhibits the synthesis of the thymine
nucleotides
5-FU converst dUMP to F-dUMP which prevents dUMP → dTMP
What is semiconservative replication?
in DNA replication: one strand is parental, the other
is a newly synthesized.
Describe the origins of replications in prokaryotes and eukaryotes
prokaryotes: replication begins at single, unique origin of replication.
eukaryotes: replication begin at several sites (AT-rich sites)
Describe the Pre-priming complex and their subunits
Unwinds double helix ahead of advancing replication fork and keep strands separated.
consist of three enzymes:
DNA helicase unwinds double helix.
DnaA protein binds and melts dsDNA.
ssDNA-binding (SSB) proteins bind to ssDNA.
keeps strand separate and protect them from nucleases
how do we prevent supercoils?
DNA topoisomerases remove supercoils and
facilitates transcription.
Describe the direction of DNA replication. Differentiate between leading strand and lagging strand
DNA polymerases read DNA sequence in 3’→5’ direction, and synthesize DNA strand in 5’→3’ direction.
Leading strand is being copied in the direction of replication fork = Synthesized continuously
Lagging strand: being copied in the direction away from the replication fork = not continuous
okazaki fragments: small frag. of DNA being created near replication fork
Describe the initiation and elongation step of DNA replication
primases are specific RNA polymerases that synthesize RNA primers
DNA pol III uses RNA primer to initiate replication. Elongation is done by this too
also major enzyme used for proofreading
For lagging strand
DNA Pol III synthesize until it is blocked by a RNA primer; then, it moves to another primer to synthesize new Okazaki fragment.
DNA Pol I replaces RNA primers with DNA.
Ligases join the ozarki fragments together
also responsible for DNA repair and recombinant DNA techniques
Ligases joins the fragments by forming phosphodiester bond between 3’ hydroxyl end of one nucleotide and 5’ phosphate end of
another nucleotide. (Requires ATP)
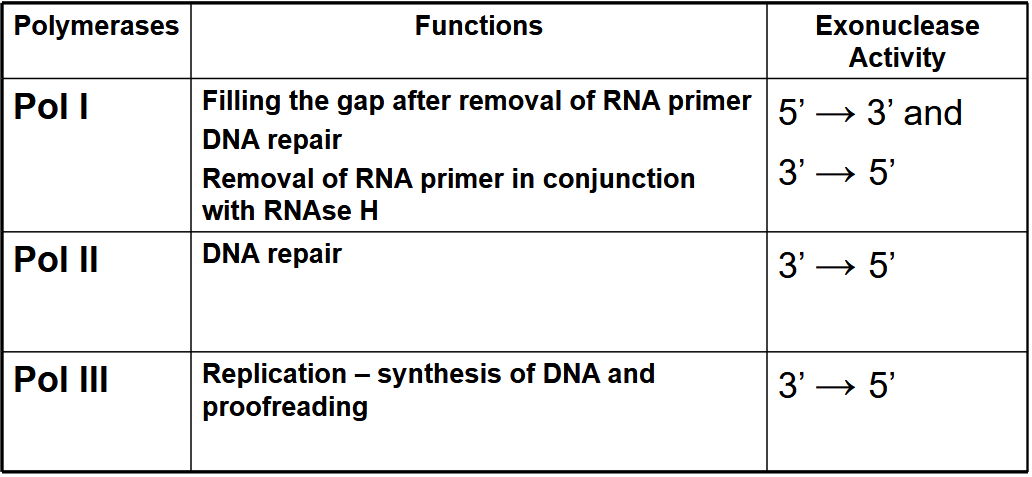
What are the functions of Pol I, Pol II and Pol III
what are the two differences in DNA replication between eukrayotes and prokaryotes
Eukaryotes
Multiple origins of replication
RNA primers are removed by RNAse H
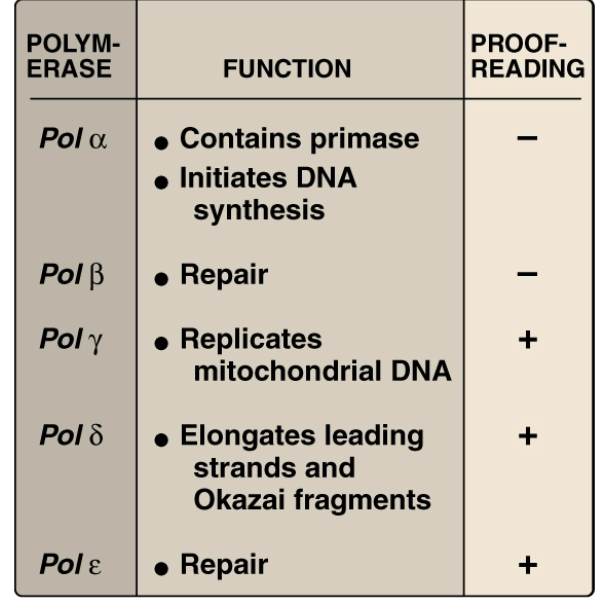
List out the different functions of the DNA polymerases used by eukaryotes
why does DNA shorten after every replication? why is this a problem?
There is no way to fill the gap after the removal of
RNA primer from extreme 5’-end of the chromosome
Consequence: loss of genetic information and is associated with aging,
What is telomeres? Their functions? What are telomerases?
at the end of DNA, contains telemers which are region of highly repetitive DNA at the end of chromosome (TTAGGG for humans)
function:
Serve as a buffer, which is consumed during cell division
Protect the ends from nucleases
Limit cells to a fixed number of divisions, in most somatic cells
Can be replenished by telomerase
Active in germ, stem, and cancer cells
How do we maintain our telomers?
excercise upregulated telomerase in aorta and mononuclear cells in mice,
Higher vitamin D level is associated with longer telomere lenght in leukocytes,
Describe the mechanism of telomerase
RNA-dependent DNA polymerase
Contains RNA which is complementary to repetitive sequence in telomere
Uses 3’ end of DNA as a primer and RNA as a template to extend 3’ end of DNA
Once a single-stranded portion of telomere end is extended, an RNA primer is produced to fill a single-stranded portion of chromosome.
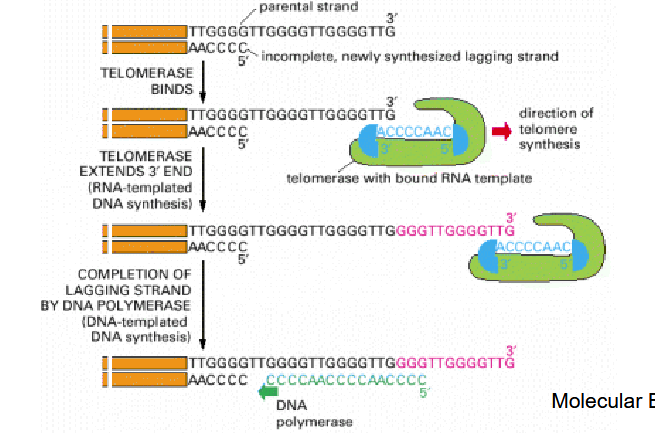
What is TERC?
Telomerase RNA component (TERC), is a component of telomerase used to extend
telomeres. TERC serves as a template for
telomere replication (reverse transcription) by
telomerase
what are some health functions disrupted by short telomers/telomerase
shorter = more hematopoisis
telomerase = a contributor to uncontrollable, indefinite growth (HeLa Cells)
what are possible causes of DNA mutations
mutagens
Chemicals produced by cells
Chemicals from environment
Radiation: X-rays and UV light
Carcinogens
mismatches; without adequate repair mech = mutation
describe how X-rays cigarettes smoke and UV light can cause DNA damage
X-Rays: produces free radicals from H2O.
cigarette smoke contains carcinogen benzo[a]pyrene
When oxidized in the body, it forms adducts with guanine residues.
UV light forms covalent dimer
between adjacent thymines on
DNA strands.
describe Xeroderma pigmentosum
Autosomal recessive
It is caused by genetic defect in nucleotide excision repair (NER) system.
Cells cannot repair damaged DNA leading to skin cancer.
Describe the four repair mechanism: direct reversal pair, NER, base excision repair, and double strand break repair
direct reversal pair:
E.coli removes thymine dimer by photactivated enzymes.
They cleave bond between bases using energy from light
nucleotide exclusion repair:
Endonuclease cleaves abnormal chain and remove distorted region.
Gap is filled by DNA polymerase.
Ends are joined by DNA ligase
Base excision repair:
DNA glycosylases recognize distortion and cleaves N-glycoside bond.
AP endonuclease cleaves sugar-phosphate strand at this site.
Same types of enzymes that are involved in other repair mechanisms finish the repair.
Double-strand break repair:
Repaired through homologous recombination.
Homologous chromosome is used as template to repair DNA.
Mutations in BRCA1 gene, a component of homologous recombination repair, result in an increased risk of breast cancer.
what four similarities connects the various repair mechanism mentioned?
Recognition of distortion.
Removal of the region with distortion.
Replacement of the gap by DNA polymerase.
Joining the ends by DNA ligase.
how does the cell know which strand is damaged and needs to be repaired?
in bacteria: only adenine of parent strand is methylated; therefore, repair is only for newly syntehsized strand
in eukaryotes: exact mechanism is not known
what is reverse transcriptase and how does viruses use this?
retroviral enzyme, uses RNA to make DNA (RNA is degraded when done)
creation of ssDNA (cDNA) is then used as the template to make dsDNA
the dsDNA can then be used for transcription of viral genes and to make copies of viral RNA.
Viral dsDNA can be incorporated
into host genome and carried from
mother to daughter cell.
what is didanosine (ddl) and explain its mechanism.
Converted to dideoxynucleoside triphosphates in cells
ddI terminates chain growth (elongation) when
incorporated into DNA by reverse transcriptase.