Exam V: Extensions, Epistasis, and Linkage
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
extension of simple medelian inheritance —> learning outcomes
LO 15.1 Distinguish between different patterns of inheritance that modify the normal Mendelian ratios: incomplete dominance and codominance, penetrance, expressivity, and lethal alleles. Relate how these different patterns result in different ratios. Describe, at a molecular level, why an individual heterozygous for a disease allele may or may not display a disease phenotype.
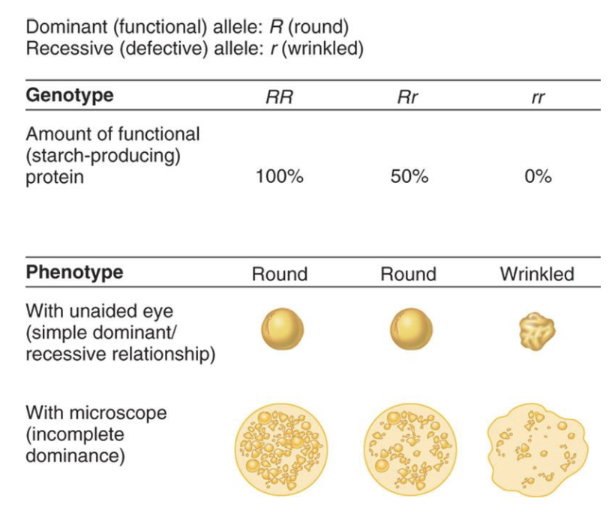
dominance/recessive at the molecular level
wild-type allele vs mutant allele
wild-type allele
the most prevalent allele in a population. encodes normal functioning protein
ex. YYRR - yellow and round are the wild types alles
mutant allele
allele that has been changed by mutation. encodes decreased or defective protein
ex. yyrr - green and wrinkled are mutant alleles
NEW mutant alleles are rare in natural populations
a mutant allele is not always detrimental
recessive allele is typically a loss-of-function mutation
there are two possible explanations for this:
50% of the normal protein is enough to accomplish the protein’s cellular function
the heterozygote may produce more than 50% of the functional protein, up regulation of a gene
wild type vs mutant
all dominant acting alleles are not the same
dominant-negative mutations
Haploinsufficiency
gain-of-function mutations
this makes it difficult to identify the causes of diseases
dominant-negative mutations
mutant proteins acts in an antagonistic manner to the normal protein
ex. p53 & uncontrolled cell growth, fibrillin-1 & marfan disease
Haploinsufficiency
dominant allele is a loss of function allele
ex. COL4A5 & alport syndrome (x-linked dominant)
gain-of-function mutations
new or abnormal function
ex. spike proteins & SARS-CoV2, HTT & Huntington’s disease
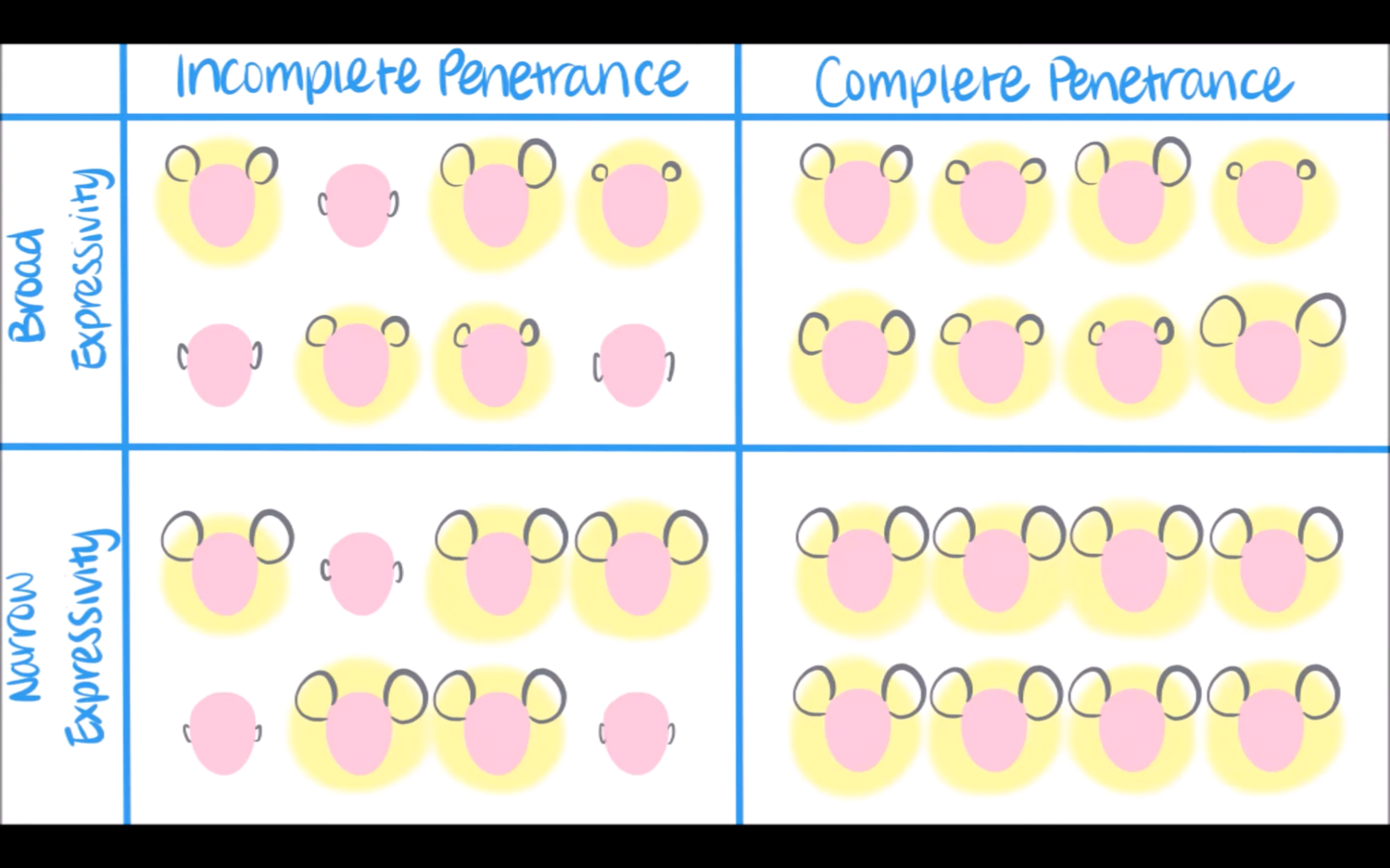
penetrance
incomplete vs complete
term indicates that a dominant allele does not always “penetrate” into the phenotype of the individual
heterozygote or carrier may or may not exhibit the mutant trait
the measure of penetrance is described at the population level
ex. if 30%, then 3 out of 10 carriers (heterozygotes) express the mutant trait
expressivity
variable or invariable
expressivity is the degree to which a trait is expressed in an individual
ex. a person with several extra digits has high expressivity. a person with a single extra digit has low expressivity
a dominant allele results in multiple digits on the hands and/or feet of humans and is determined to be 50% penetrant. this means that:
A. 50% of the individuals in the population will exhibit the trait.
B. 50% of the homozygous dominant individuals in the population will exhibit the trait.
C. 50% of the heterozygotes in the population will exhibit the trait.
D. 50% of the individuals exhibit varying levels of trait expression but the trait is always present in an individual.
C. 50% of the heterozygotes in the population will exhibit the trait.
heterozygotes mean “carriers” and that is what penetrance deals with carriers
what are the different types of domiance?
lethal alleles
incomplete dominance
codominance
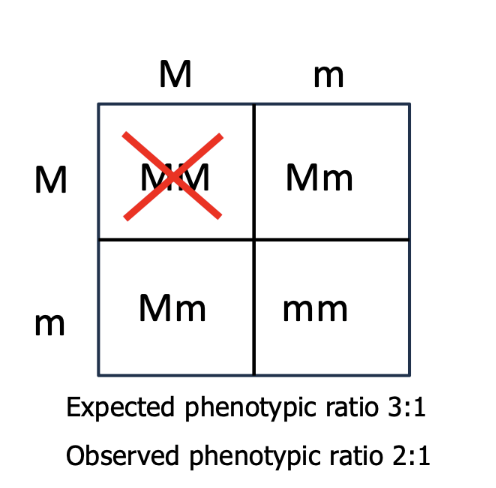
lethal alleles
complete dominance & lethality
an ex. can be seen through manx cats
tailless phenotype is caused by a dominant allele
two copies of the dominant allele, for the tailless allele, is lethal and results in the death of the embryo
therefore, you can never have a true-breeding tailless cat since you can never be homozygous dominant for a dominant lethal allele
expect phenotype is 3:1 but due to lethal allele types the observed phenotypic ratio is only 2:1
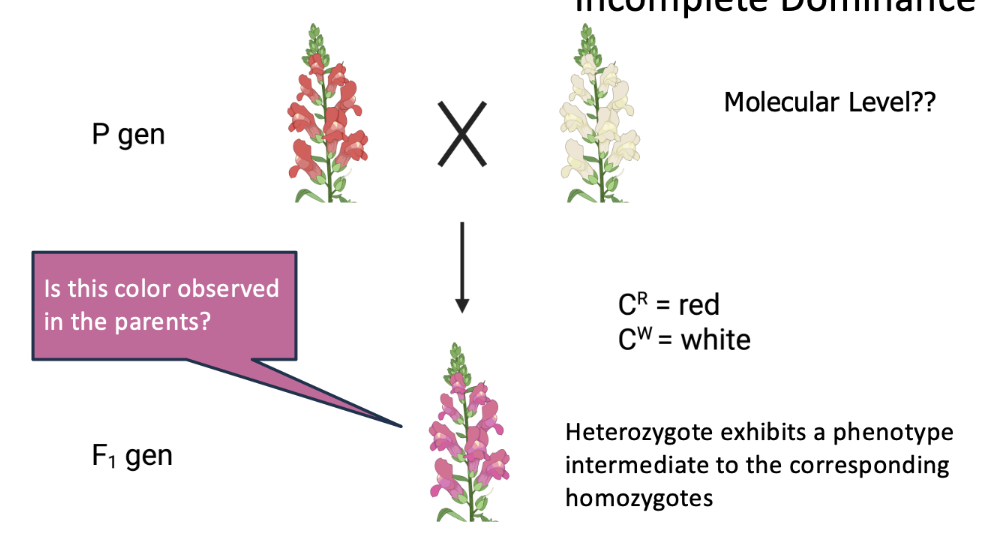
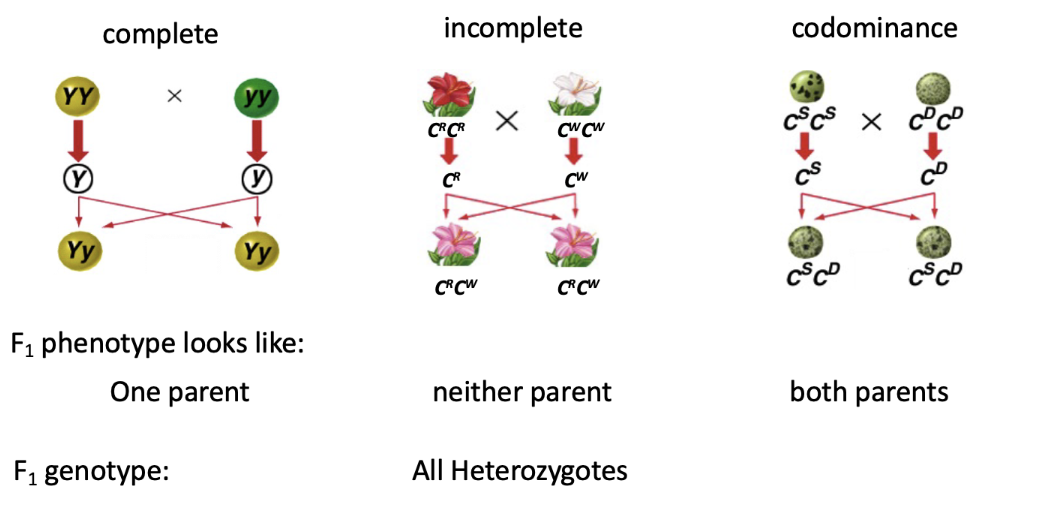
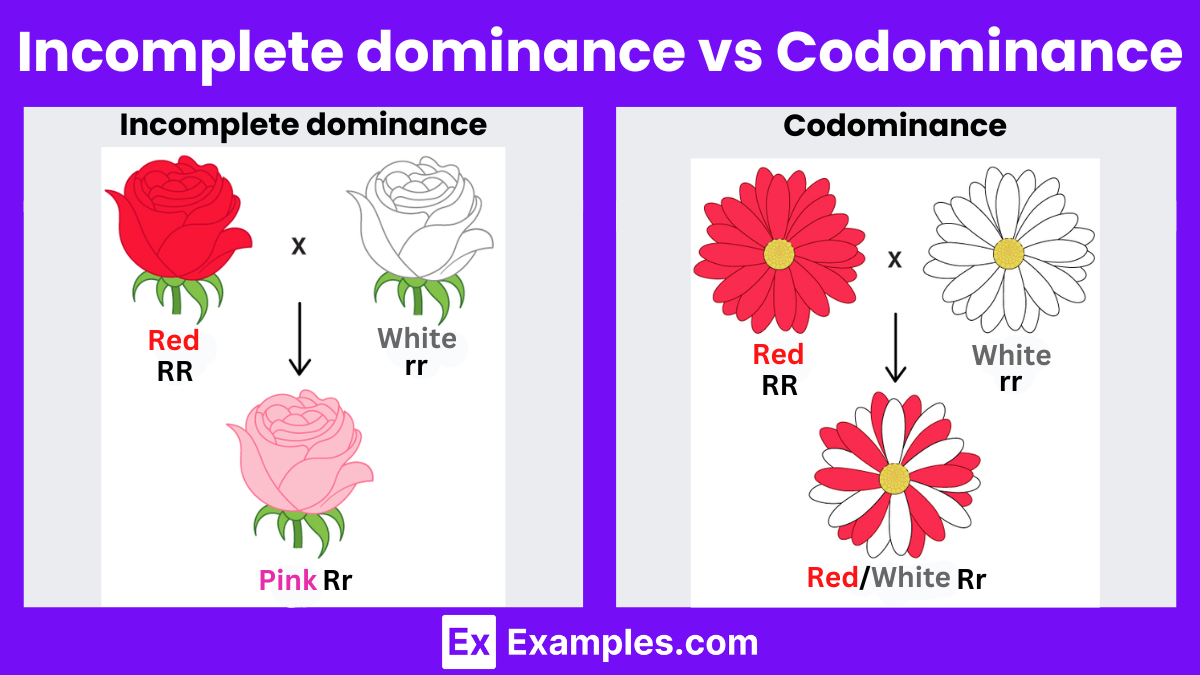
incomplete dominance
type of genetic inheritance where the heterozygous phenotype is a blend of the two parental traits, rather than one being completely dominant over the other.
In simpler terms, neither allele is fully dominant, so the resulting trait in the offspring is a mix of both.
In snapdragon flowers:
A red-flowered plant (RR) crossed with a white-flowered plant (WW) produces pink flowers (RW).
The pink color is a blending of red and white, showing incomplete dominance.
It's different from complete dominance (where one trait fully masks the other) and codominance (where both traits show up equally, like in AB blood type).
where a heterozygote exhibits a phenotype intermediate to the corresponding homozygotes
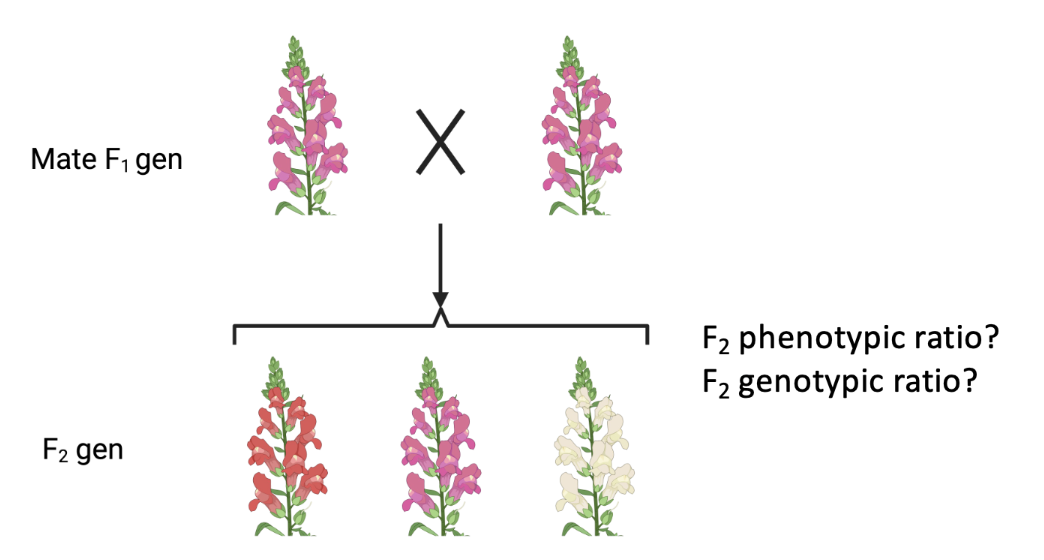
think of it as the absence of red is white
codominance
a type of genetic inheritance where both alleles in a heterozygous organism are fully and equally expressed. Instead of blending, both traits appear side by side in the phenotype.
see both traits clearly
In human blood types:
A person with one A allele and one B allele has AB blood type.
Both A and B antigens are equally expressed—neither is dominant or recessive.
Another example is in roan cattle:
A red cow (RR) crossed with a white cow (WW) produces offspring with red and white hairs intermixed (RW)—not pink (incomplete domiance), but both colors visible.
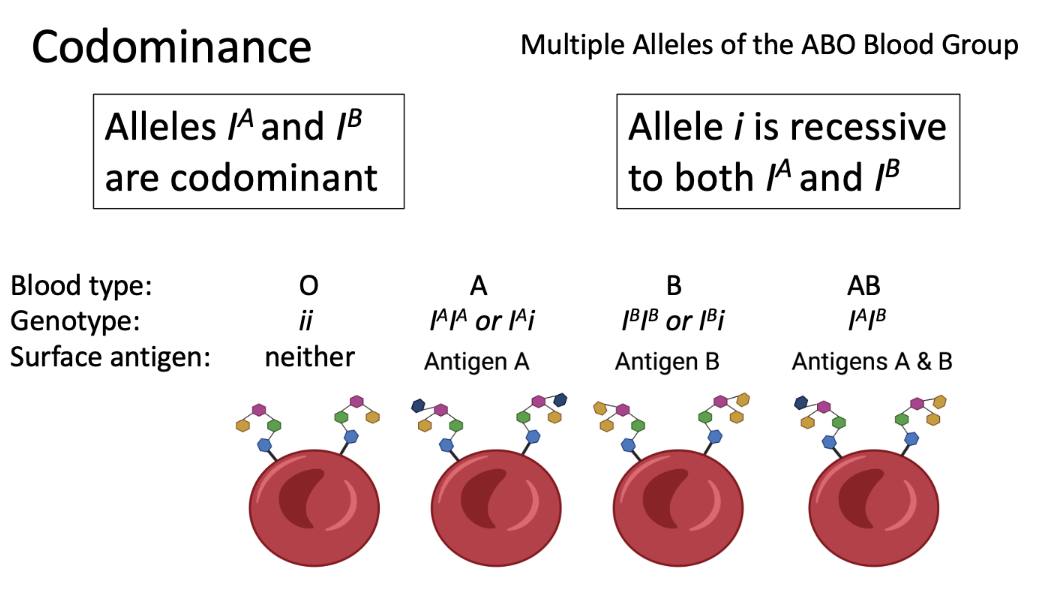
universal donor
universal acceptor
universal donor: O ii
universal acceptor: AB IAIB
summary for the 3 types of dominance
the blood type alleles in humans show example(s) of which type(s) of inheritance patterns?
codominance
complete dominance
multiple alleles
lethal alleles
incomplete dominance
codominance
complete dominance
multiple alleles
Keritinosis is a recessive disorder that causes dry skin on hands and feet. Use keritinosis to provide an example of:
A) variable expressivity
B) incomplete penetrance.
A) Variable expressivity
Some people with a kk genotype have dry hands and some have dry feet, some have dry hands and feet
B) Incomplete penetrance
Not everyone with kk genotype has a dry skin phenotype
polygenic & complex traits, gene interactions learning outcomes
LO 16.1 Explain, and illustrate with examples, the differences in phenotypic variation when traits are due to genetics only, environment only, and genes and environment together.
LO 16.2 Using an additive model of allele contributions to a trait, determine the phenotypes of individuals who have varying numbers of dominant alleles for a polygenic trait.
LO 17.1 Describe the phenomenon of epistasis and interpret the results of crosses involving genes working in a pathway. Given a set of genes that are known to interact in a pathway, determine the “epistatic” relationship between the genes. Predict phenotypes of offspring of crosses involving two genes, given information about biochemical pathways or functions of the genes.
LO 17.2 Explain the rationale of a complementation test and interpret the experimental results of such tests.
phenotypes can be determined by…
genetics (ex. person with PKU)
environment
genetics + environment (ex. simese cats who have temperature dependent alleles in fur color)
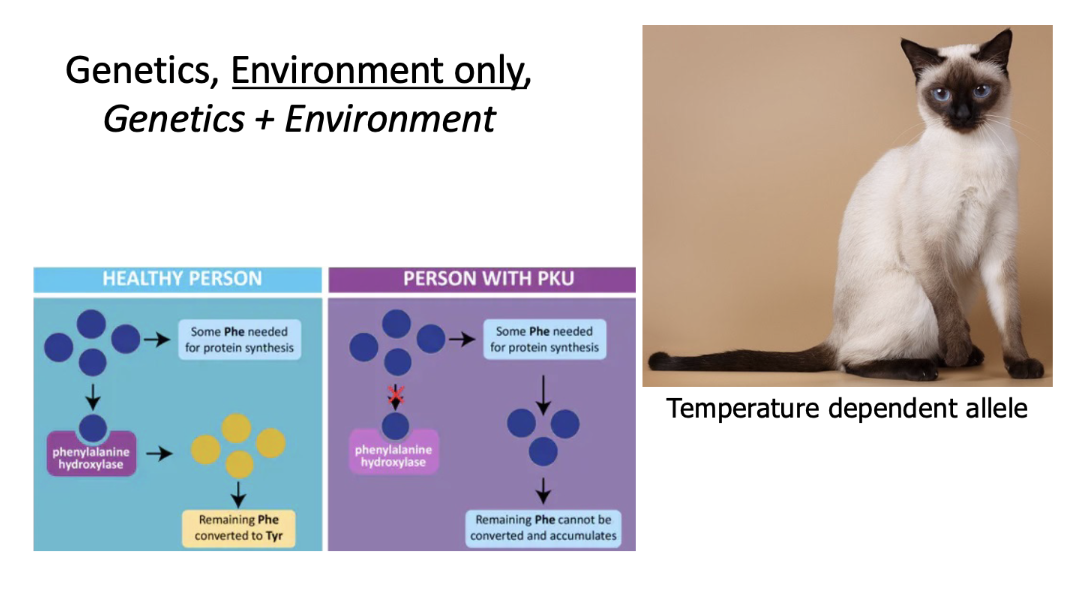
simese cats: darker colored extremities in cooler regions - have higher amounts of melanin (combo of environmental and genetics)
polygenic characters
multiple genes are involved in producing phenotype, ex. skin color
continuous traits meaning there are no discrete phenotypic classes
additive effects of DOMINANT alleles
trihybrid cross showing polygenic traits in skin color
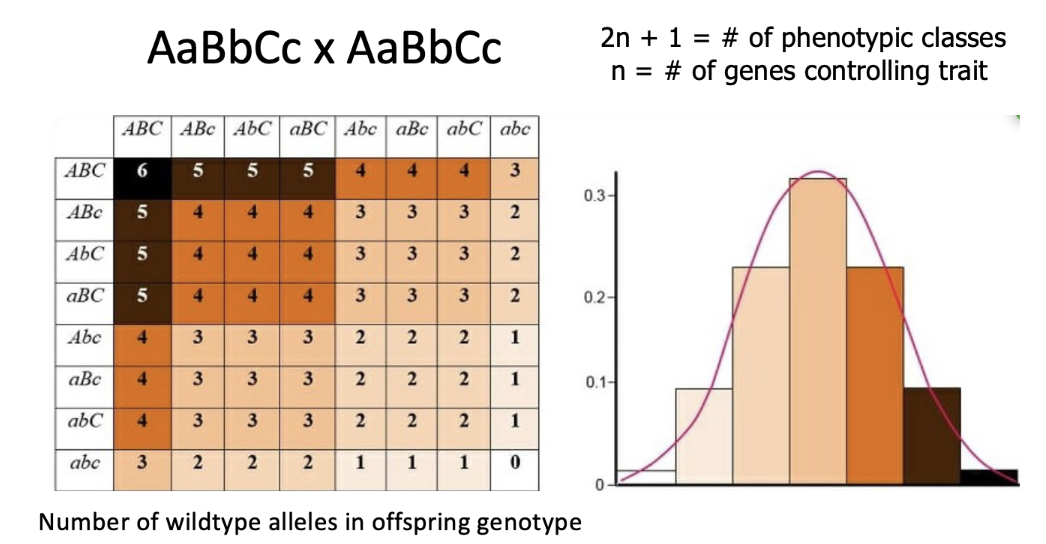
increase in # of dominant alleles —> darker spectrum
continuous characteristics
how do you find the # of phenotypic classes? # of genes controlling the trait in polygenic characters?
# of phenotypic classes - 2n+1
# of genes controlling the trait - n
gene interactions
when two or more different genes influence the outcome of a single trait
examples include:
complementation
epistasis
ex. sweet pea plant
complementation
each have one copy of a working allele

in generation I —> AAbb x aaBB
mutations are found in two different genes, gene A and gene B, represented by alleles “a” or “b”, causing both parents to be deaf
in generation II —> AaBb
due to the complementation of the genes, there is one working allele “A” or “B” in each offspring so none of the children are deaf
non complementation
mutation is found in only one gene (failure to complement)

in generation I —> AAbb x AAbb (or aaBB x aaBB)
mutations are found in only one gene of each parent, either gene A or gene B, represented by alleles “a” or “b”, causing both parents to be deaf but because of the same allele mutation
in generation II —> AAbb (or aaBB)
due to the non complementation of the genes, there is no working alleles so all of the children are deaf
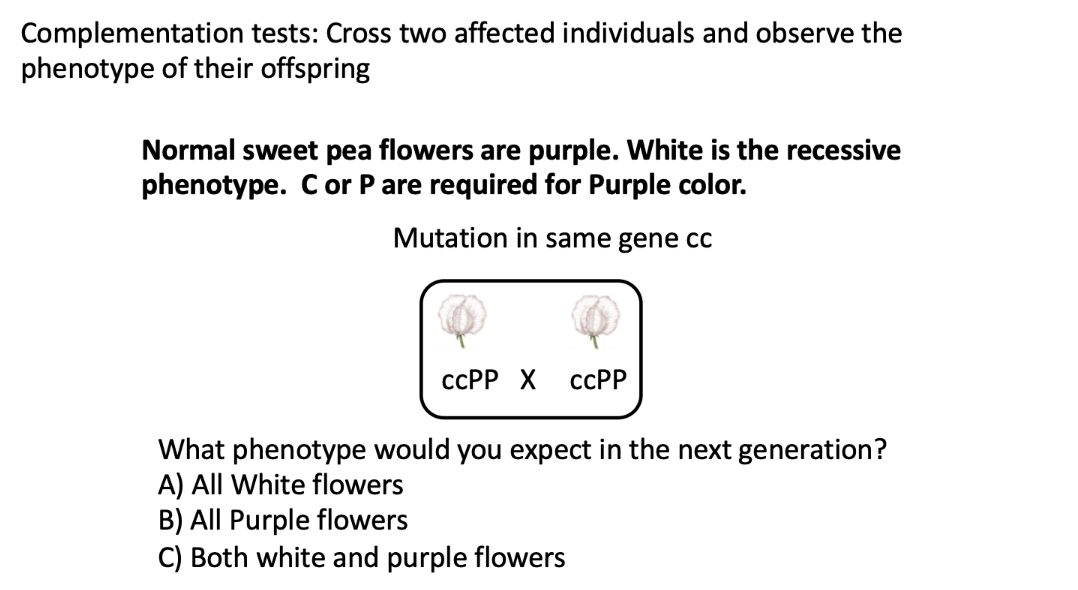
A) All white flowers
both parents are mutant in the same gene
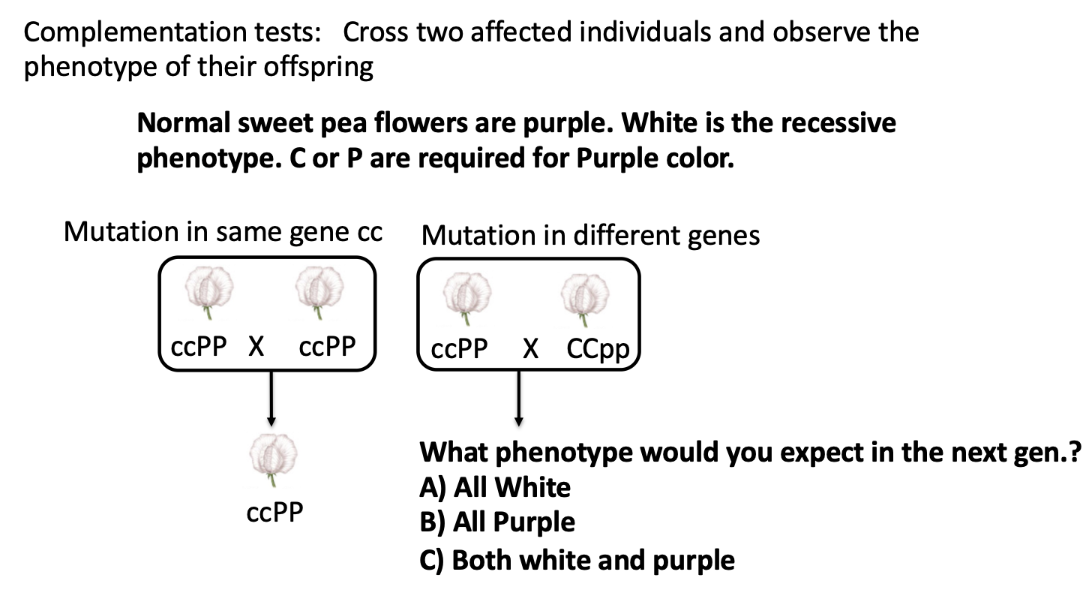
B) All Purple
complementation —> all heterozygous offspring with one working allele in each gene
break down of mutations
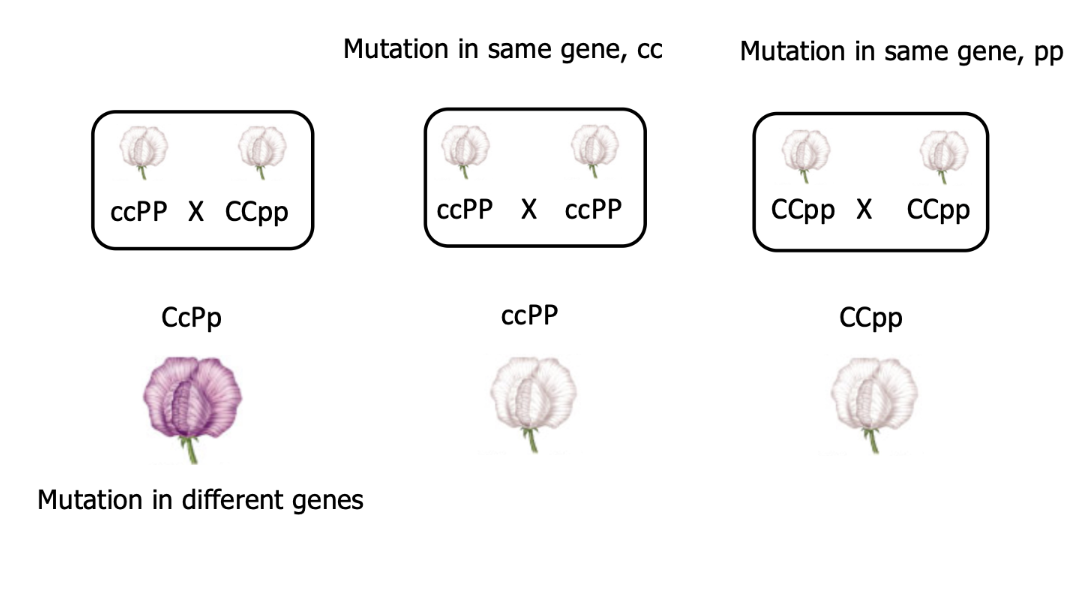
how can we determine how many genes are involved in a single phenotype?
you expose wild type purple flowers to Ultraviolet radiation to see if you can identify a bunch of new mutations and determine the number of genes involved in flower color.
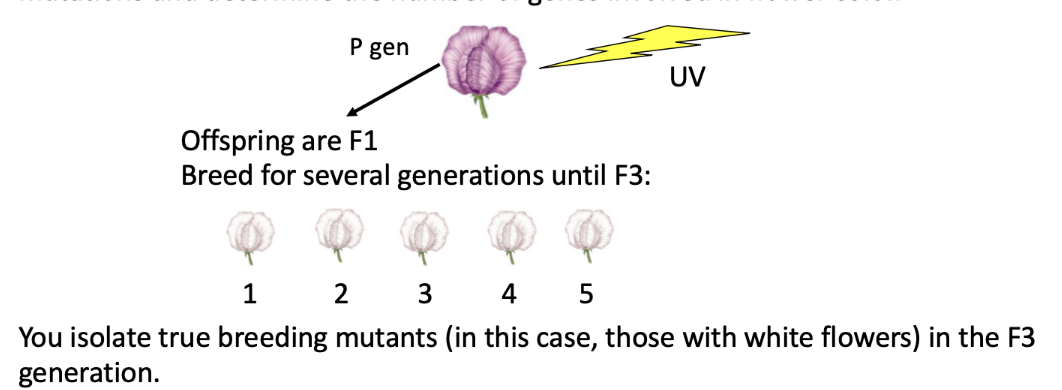
all of these flowers are white because of the same mutation!
then you set up a complementation test
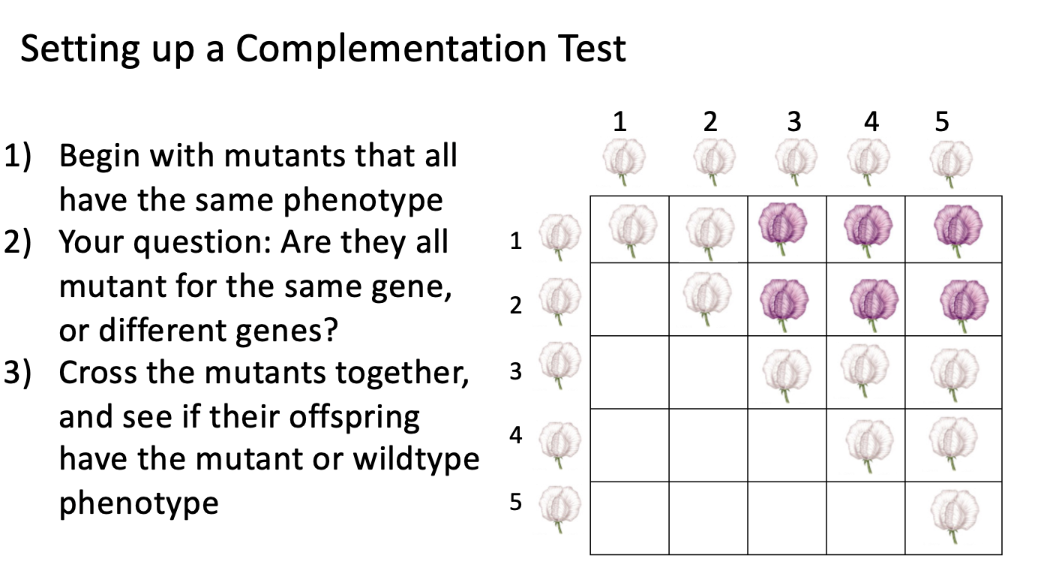
after setting up the complementation test…
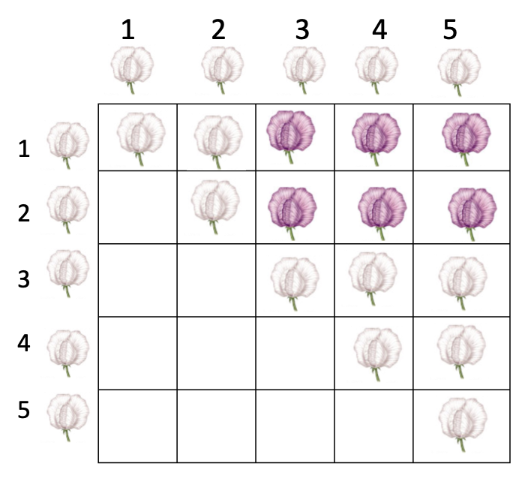
no complementation (-)
complementation (+)
no complementation
(-)
mutations in the same gene, contributing to a white flower!
complementation
(+)
mutations in different genes, wild type flower (purple)!
complementation group
when two mutations don’t complement each other, mutations in same gene
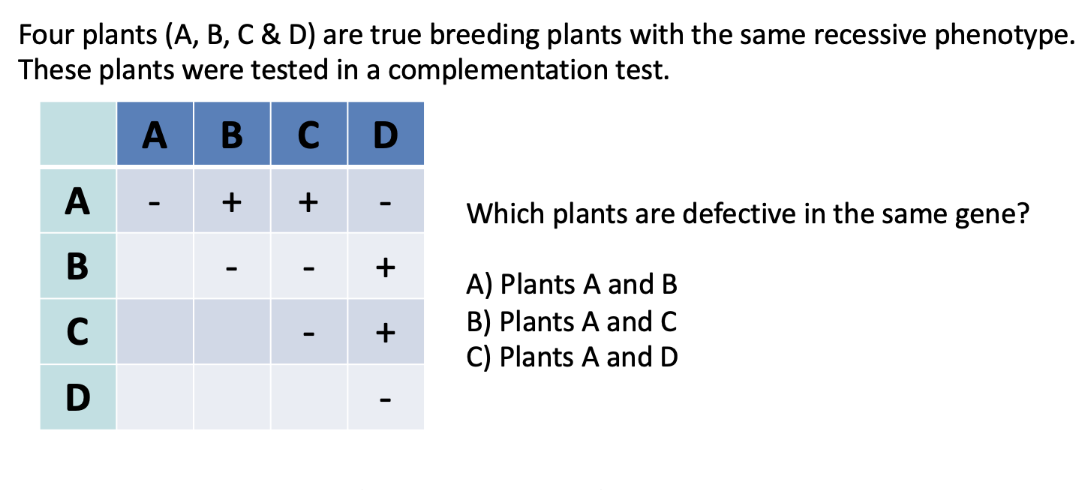
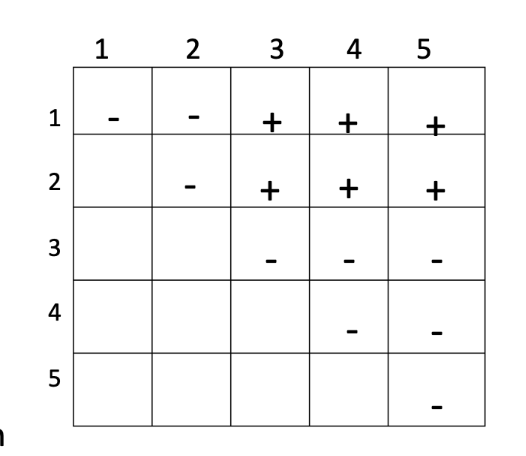
ANSWER: C (plants A and D)
+ = complementation (normal phenotype)
- = no complementation (mutant phenotype)
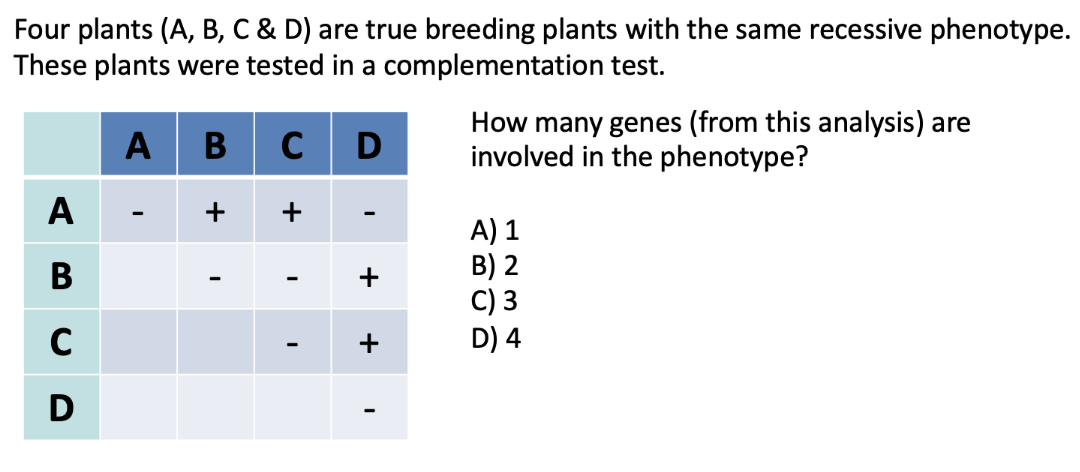
ANSWER: B (2)
+ = complementation (normal phenotype)
- = no complementation (mutant phenotype)
sometimes genes code for protein products working sequentially in a pathway to establish a particular phenotype
in such a case, mutations in the genes lead to phenotypes with an unexpected ratio
epistasis
when alleles of one gene mask the phenotypic effects of the alleles of another
two different genes interact together to affect the SAME trait
epistatic gene - the gene whose expression interferes with or masks the effects of another gene
hypostatic gene - the gene whose expression is suppressed
mendel’s laws of segregation and independent assortment still apply
ratios in F2 offspring are variations of the 9:3:3:1 ratio
9:4:3 or 12:3:1
recessive epistasis
phenotypic ratio 9:3:4
when two (or more) genes can be involved in producing a particular phenotype
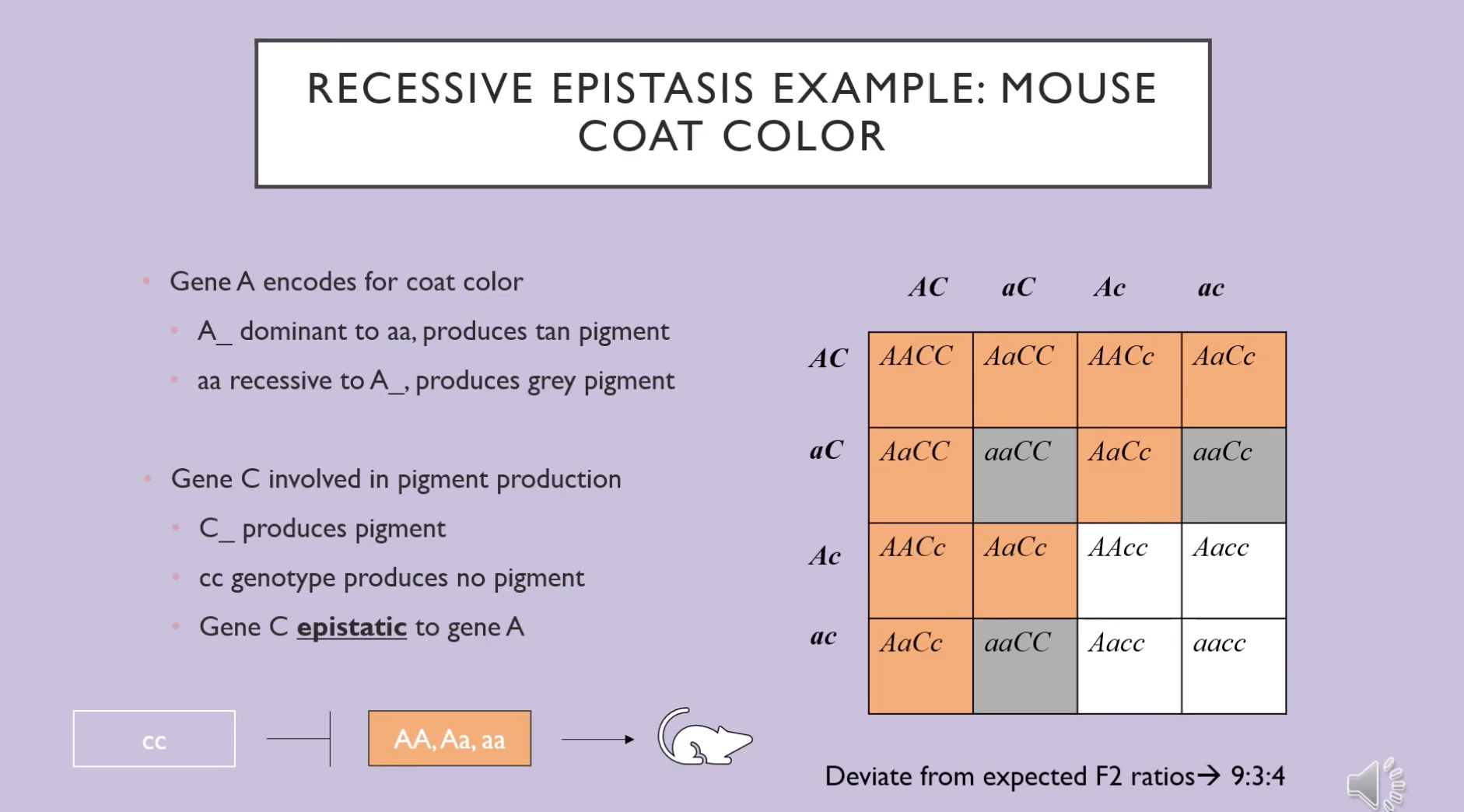
example of recessive epistasis
labs

dominant epistasis
dominant allele at locus A means no pigment produced. Gene A is epistatic to gene B
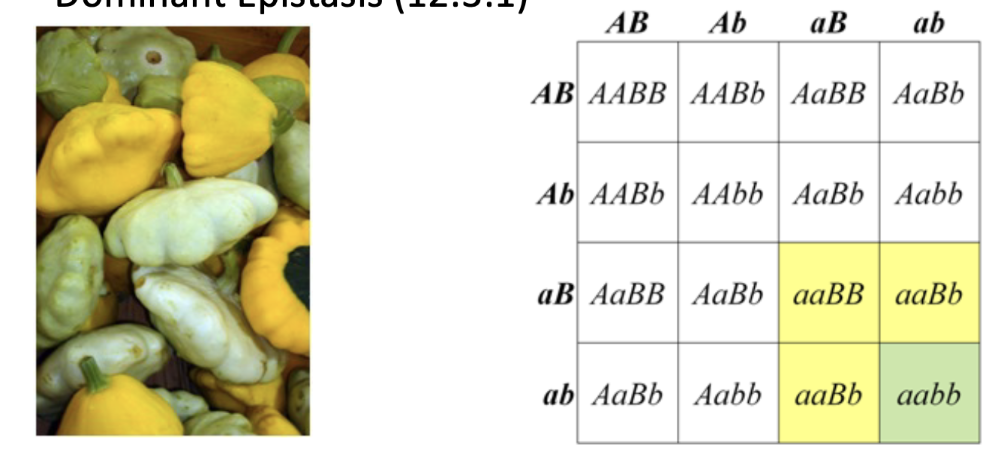
observed 12:3:1 phenotypic ratio
dominant allele of one gene hides the expression of the alleles of another.
ex. dominant A allele is masking the genes of the B or b allele.
gene A is epistatic to gene B
incomplete dominance vs codominance
penetrance vs expressivity
penetrance: how often does someone with the genotype actually show the corresponding
expressivity: variation in severity or expression of phenotype
genetic linkage and recombination
LO 18.1 Explain how homologous recombination during meiosis leads to new combinations of alleles in gametes, at different proportions than with unlinked genes.
LO 18.2 Determine haplotypes, genetic map distances, and recombination frequencies by analyzing progeny with recombinant phenotypes from genetic crosses. Understand that linked genes do not exhibit independent assortment because recombination will not always occur between the loci. Distinguish between parental and recombinant chromosomes, gametes, and offspring, and identify them in crosses. Calculate the map distance between loci given the phenotypes of offspring or predict phenotypes of offspring given the recombination frequency between loci. Use the distance to construct genetic maps based on data from two-point testcrosses.
LO 18.3 Calculate the probability of a particular gamete being produced from an individual when provided with the map distance between two linked genes.

E) All of the above
A) doesn’t tell us a lot, other than the alleles presented
B) and C) show more information such as were the alleles are located
linkage can be…
physical or genetic
synteny
two or more genes are located on the same chromosome and are physically linked
they may or may not be genetically linked
genetic linkage
the phenomenon that genes close together on a chromosome tend to be transmitted as a unit
can disrupt expectation of based on independent assortment
all genetically linked genes are syntenic but not all syntenic genes are genetically linked
example of a linkage map for Drosophila melanogaster

where the outlined in red are not genetically linked or even synteny. independent assortment will occur
purple are syntenic - since they are located on the same chromosome
green are both syntenic and genetically linked
numbers on the side mark the locations of genes. provide you with a general idea of the location and distance apart

B) More AB gametes than Ab gametes
A) expectation of independent assortment but since they are genetically linked, they won’t be experiencing that
B) Ab gametes would be few and far between but possible bc of recombination occuring due to crossing over
C) not possible to have more recombinants than to have of nonrecombinants
D) can not be expected because we know that recombination will occur
assuming complete interference —> no crossing over whats so ever
unlinked genes assort independently but genes on the same chromosome do not, but…
alleles still segregate, but….
what are the haplotypes of the gametes?
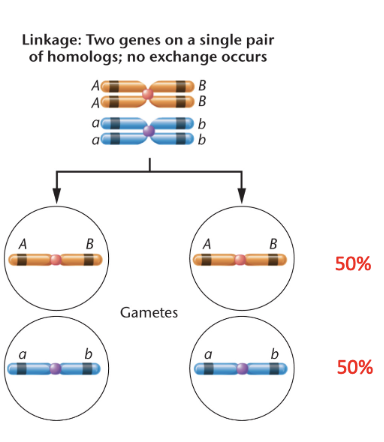
haplotypes of the gametes
AB
ab
define haplotypes: what alleles are presented in the gametes
the inheritance of linked alleles occurs at a higher frequency than those of unliked alleles
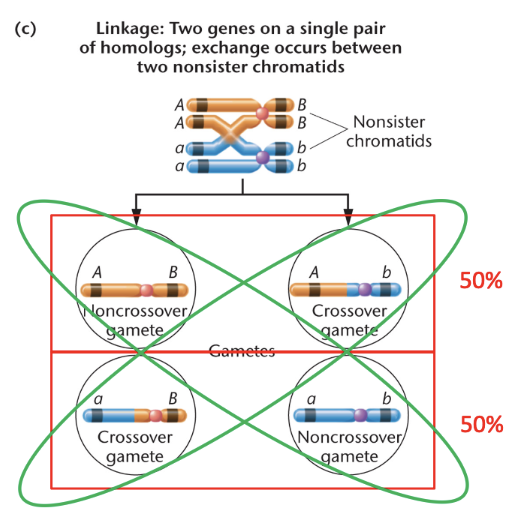
recombination via crossing over REMINDER
recombination refers to the creation of new allele combinations on homologous chromosomes
crossing over occurs during prophase of meiosis I but it doesn’t always result in new combinations of alleles
only get new allele combinations with crossing over between non-sister chromatids
can not occur in sister chromatids because they are identical and you won’t see any change in the genotypes or the phenotype
crossing over between sister chromatids will not ever occur because they are identical
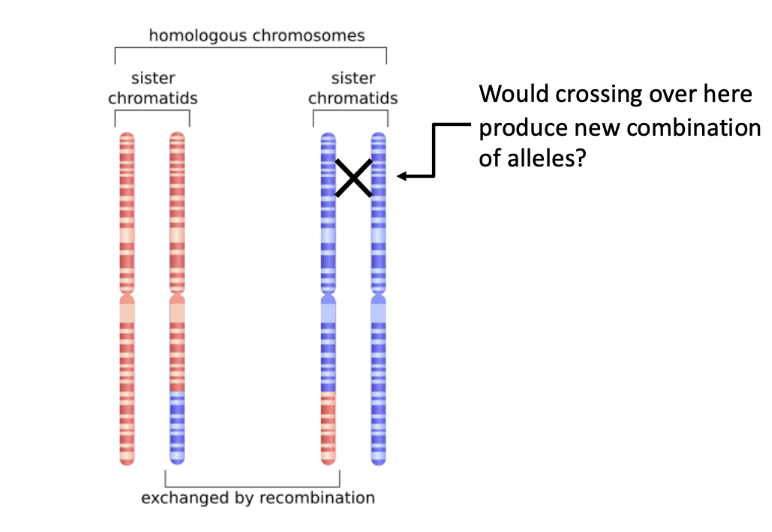
how can you test to determine if these genes are linked
assuming that traits are produced by two genes with two variants each
complete dominance, penetrance, and no variable expressivity
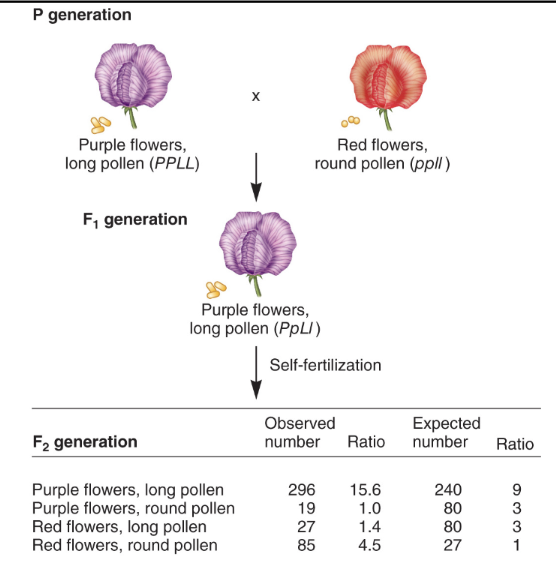
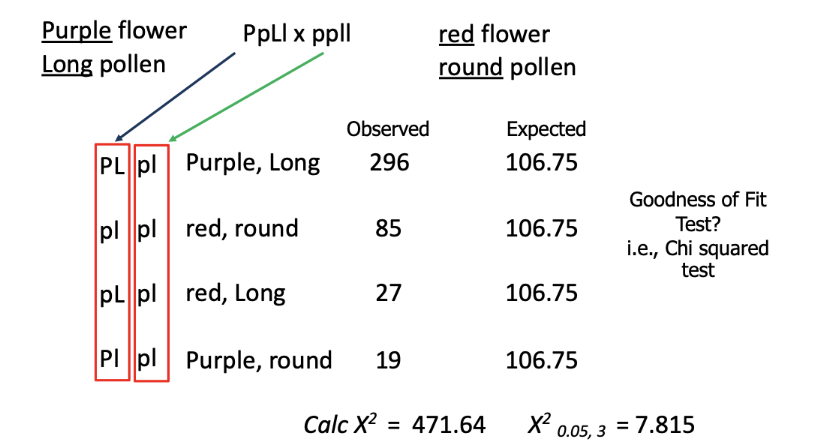
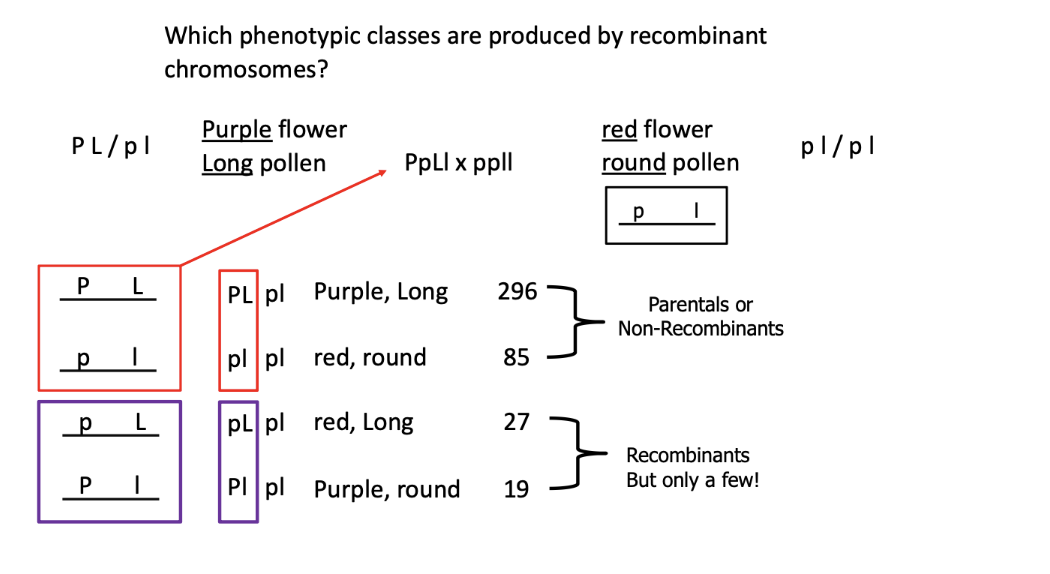
dihybrid test cross to determine if these genes are linked.
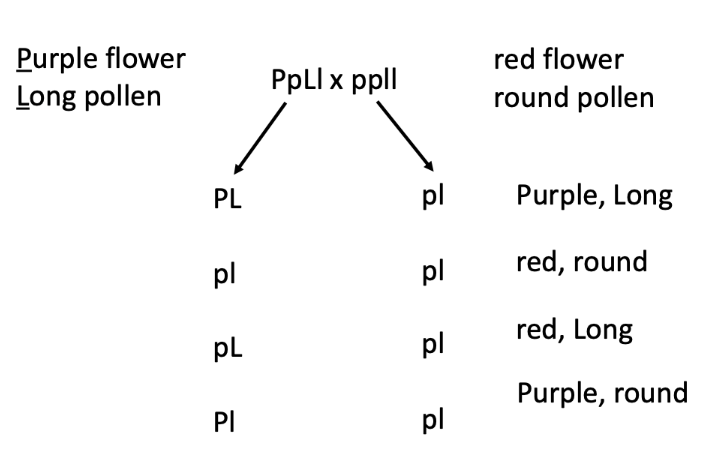
null hypothesis: if independent assortment, then would expect a 1:1:1:1 ratio in the offspring when conducting a dihybrid test cross
what is a test cross
when you mate a double heterozygous w/ a double homozygous recessive
what phenotypic classes are produced by recombinant chromosomes?

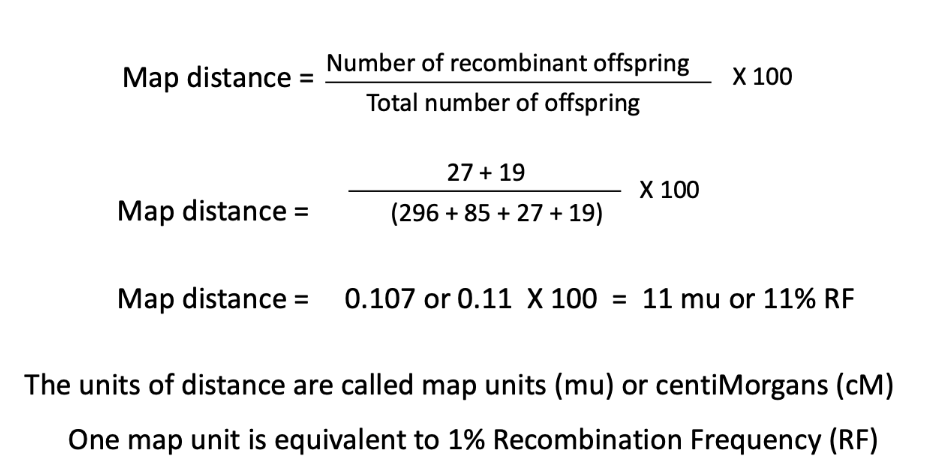
how do i calculate the map distance between two genes?
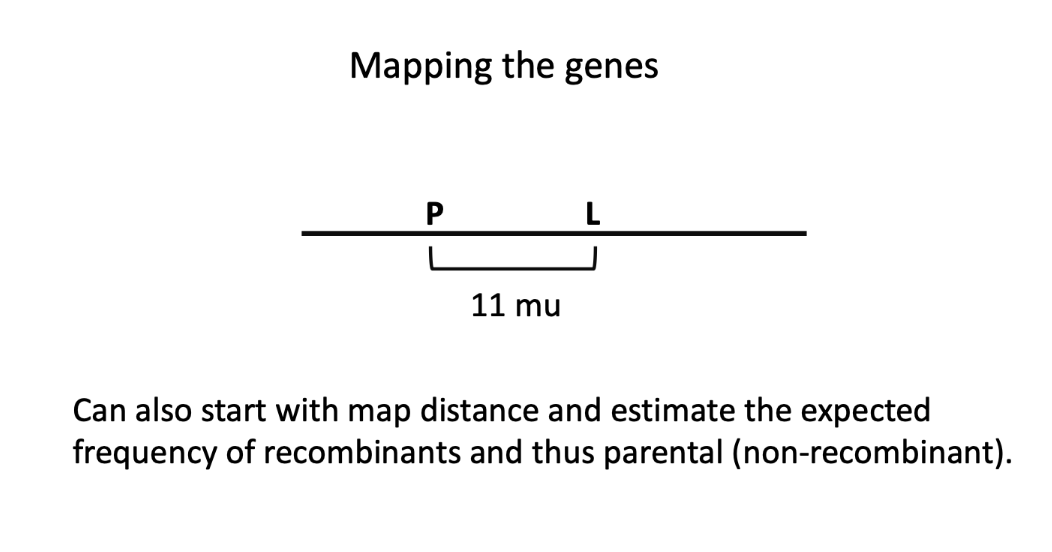
if genes are syntenic and > 50 mu apart
then the genes independently assort
if genes are syntenic and < 50 mu apart
genetically linked
ONLY GENETIC LINKAGE: percentage of recombinant offspring is a function of the distance between the two genes
if the genes are far apart (but <50 mu) then there will be many recombinant offspring
as compared to 10 mu which would have fewer recombinant offspring
if the genes are close then there will be fewer recombinant offspring