Cell and Molecular Biology Quizzes
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
45 Terms
C) Cells in culture grow in a rigid 2D substrate and are not exposed to the same array of biochemical and mechanical signals they would encounter in vivo.
Which of the following is a key difference between the conventional cell culture environment and the physiological conditions in the body?
A) Cells in culture receive the same variety of signals from growth factors and cytokines as they do in the body.
B) Cells in culture are exposed to a complex 3D structure similar to their native environment.
C) Cells in culture grow in a rigid 2D substrate and are not exposed to the same array of biochemical and mechanical signals they would encounter in vivo.
D) Cells in culture interact with the immune system in the same way they do in the body.
B) They are easier to standardize, involve stem cells or multiple cell types, and allow controlled study of cell-ECM interactions.
What is a key benefit of using 3D culture systems such as organoids or spheroids over native tissue for studying biological systems?
A) They offer more complex tissue architecture and are harder to reproduce across different experiments.
B) They are easier to standardize, involve stem cells or multiple cell types, and allow controlled study of cell-ECM interactions.
C) They completely replicate the in vivo environment without the need for external control.
D) They do not require stem cells and are simpler than native tissues in terms of biological complexity.
B) When the goal is to determine toxicity, investigate internal cellular processes, or purify specific molecules in a simplified environment.
When are in vitro systems generally preferred over in vivo systems in scientific research?
A) When studying complex interactions among multiple cell types and tissues over long time scales.
B) When the goal is to determine toxicity, investigate internal cellular processes, or purify specific molecules in a simplified environment.
C) When assessing the biocompatibility of new materials in the physiological environment.
D) When investigating physiological mechanisms that require the interaction of inflammatory cells.
A) Primary cultures retain many native cell characteristics, making them valuable for studying native cell behavior, but they have a finite lifespan and more challenging culture conditions. Established cell lines, in contrast, are easier to grow and manipulate but may lose some native characteristics over time.
Which of the following statements best describes the advantages and limitations of primary cultures versus established cell lines?
A) Primary cultures retain many native cell characteristics, making them valuable for studying native cell behavior, but they have a finite lifespan and more challenging culture conditions. Established cell lines, in contrast, are easier to grow and manipulate but may lose some native characteristics over time.
B) Primary cultures are easier to grow and manipulate, while established cell lines are less consistent and harder to maintain.
C) Primary cultures are preferred for high-throughput experiments, while established cell lines are used for studying specific molecular pathways.
D) Established cell lines always retain the original characteristics of the cells and are ideal for studying native cell behavior.
C) Scanning Electron Microscopy (SEM)
Which of the following microscopy techniques provides high-resolution, 3D images of the surface structures of cells and tissues?
A) Bright-field Microscopy
B) Phase Contrast Microscopy
C) Scanning Electron Microscopy (SEM)
D) Transmission Electron Microscopy (TEM)
D) Cell typing focuses on classifying cells based on molecular markers or genetic expression, while cell phenotyping focuses on assessing observable traits and functional behaviors of the cell.
Which of the following best differentiates cell typing from cell phenotyping?
A) Cell typing identifies the functional activity of a cell, while cell phenotyping classifies a cell based on its identity and origin.
B) Cell phenotyping classifies cells by their developmental lineage, while cell typing examines the genetic profile of the cell.
C) Cell typing is used primarily for studying immune cells, while cell phenotyping is used for examining tissue-specific cells only.
D) Cell typing focuses on classifying cells based on molecular markers or genetic expression, while cell phenotyping focuses on assessing observable traits and functional behaviors of the cell.
A) Dissect tissue layers, digest the extracellular matrix (ECM) to liberate the cells, and use flow cytometry to separate the specific cell types.
Which of the following methods would a biomedical engineer use to isolate a pure population of arterial cells for culture?
A) Dissect tissue layers, digest the extracellular matrix (ECM) to liberate the cells, and use flow cytometry to separate the specific cell types.
B) Collect the entire tissue and culture it without any separation of cell types.
C) Isolate only endothelial cells using a rigid substrate in culture without additional treatments.
D) Culture smooth muscle cells and fibroblasts together within the ECM, without isolating them first.
B) Western Blotting
Which of the following techniques is used to detect specific proteins in a sample by separating proteins through gel electrophoresis and probing with antibodies?
A) Flow Cytometry
B) Western Blotting
C) RNA Sequencing (RNA-Seq)
D) Chromatin Immunoprecipitation (ChIP)
D) To promote cell behaviors, such as adhesion, migration, and differentiation, that are more similar to the natural tissue environment.
Why might a biomedical researcher choose to coat the culture surface with components of the extracellular matrix (ECM) such as collagen, fibronectin, or laminin?
A) To create a more artificial environment that suppresses cell adhesion and differentiation.
B) To eliminate the need for any cell-cell interactions in the culture system.
C) To simplify the culture process by preventing cells from adhering to the surface.
D) To promote cell behaviors, such as adhesion, migration, and differentiation, that are more similar to the natural tissue environment.
C) To differentiate between various tissue types and cell components, and to assess tissue morphology and abnormalities.
What is the primary use of Hematoxylin and Eosin (H&E) staining in histology?
A) To identify the specific genetic profile of a tissue sample.
B) To visualize the function of intracellular signaling pathways.
C) To differentiate between various tissue types and cell components, and to assess tissue morphology and abnormalities.
D) To label only the cell membrane for surface protein analysis.
A) The transcriptome is the complete set of RNA molecules transcribed from the DNA, including both coding and non-coding RNAs, and is dynamic and cell-specific.
Which of the following statements best describes the transcriptome?
A) The transcriptome is the complete set of RNA molecules transcribed from the DNA, including both coding and non-coding RNAs, and is dynamic and cell-specific.
B) The transcriptome is the complete set of all DNA sequences in a cell.
C) The transcriptome refers to all proteins expressed in a cell at any given time.
D) The transcriptome is a fixed collection of RNA molecules that do not change in response to environmental factors.
D) Allows real-time tracking of proteins in live cells and tissues
What is the most significant advantage of protein tagging over antibody labeling?
A) Higher specificity and reduced background signal
B) Can detect a wider range of molecules
C) Does not require any fluorescent dyes
D) Allows real-time tracking of proteins in live cells and tissues
B) To amplify the signal by binding to the primary antibody
What is the purpose of using a secondary antibody in fluorescence microscopy?
A) To directly bind to the target protein
B) To amplify the signal by binding to the primary antibody
C) To remove non-specific binding
D) To bind to the sample's DNA
C) That the two proteins are located in the same area of the sample
What does the co-localization of red and green signals in a merged fluorescence image typically indicate?
A) The complete absence of a specific protein
B) That the signals come from separate regions in the sample
C) That the two proteins are located in the same area of the sample
D) That the fluorescence has faded due to photobleaching
B) Allows for high-resolution, whole slide imaging with automation
What is the main benefit of using a slide scanner over a conventional microscope in fluorescence microscopy?
A) Provides live-cell imaging capabilities
B) Allows for high-resolution, whole slide imaging with automation
C) Offers real-time manipulation of the sample
D) Requires less expertise to handle multiple fluorophores
B) They reduce non-specific binding and offer higher specificity
Which of the following is an advantage of using monoclonal antibodies in fluorescence microscopy?
A) They can recognize multiple epitopes on the same antigen
B) They reduce non-specific binding and offer higher specificity
C) They bind to a broader range of proteins
D) They are easier to label with fluorophores
A) The fading of fluorescence over time, known as photobleaching
What is one of the challenges associated with fluorescence microscopy?
A) The fading of fluorescence over time, known as photobleaching
B) The inability to observe protein localization in live cells
C) The difficulty of labeling specific molecules in the sample
D) The lack of specialized equipment needed for imaging
D) The secondary antibody only
In fluorescence microscopy, what would the negative control condition likely consist of when using antibody labeling?
A) A sample without any antibodies
B) A sample with both primary and secondary antibodies
C) A sample treated with a different fluorophore
D) The secondary antibody only
B) It rejects out-of-focus light, ensuring that only light from the focal plane is captured
Which of the following best describes the role of the pinhole in confocal microscopy?
A) It allows light from all focal planes to pass through for a complete image
B) It rejects out-of-focus light, ensuring that only light from the focal plane is captured
C) It helps to generate 3D images by scanning the entire specimen
D) It enhances fluorescence signal intensity by concentrating light
C) The quality of the original image limits the effectiveness of the computational correction
What is the primary limitation of deconvolution imaging?
A) It cannot generate high-resolution 3D images
B) It requires the use of a confocal microscope
C) The quality of the original image limits the effectiveness of the computational correction
A) It provides high-resolution 3D imaging with minimal photodamage
What is the key advantage of using confocal microscopy for live-cell imaging?
A) It provides high-resolution 3D imaging with minimal photodamage
B) It allows for the simultaneous imaging of multiple samples
C) It can only be used with fixed, non-living cells
D) It is faster than standard fluorescence microscopy
D) By adding or removing electrons to give the droplets an electrical charge
In a FACS system, how are droplets containing single cells separated from those without cells?
A) By using a laser that burns the droplets containing cells
B) By using a physical filter that only allows droplets containing cells to pass through
C) By measuring the scatter of light from the droplets
D) By adding or removing electrons to give the droplets an electrical charge
A) Lamin is primarily localized at the nuclear membrane, forming the nuclear lamina, which provides structural support to the nuclear envelope and is involved in regulating DNA organization.
Examine the fluorescence image of cells labeled with lamin (green), tubulin (red), and DNA (blue). Based on the image, which of the following best describes the localization and function of lamin in these cells?
A) Lamin is primarily localized at the nuclear membrane, forming the nuclear lamina, which provides structural support to the nuclear envelope and is involved in regulating DNA organization.
B) Lamin is present throughout the cytoplasm, where it associates with microtubules to stabilize cellular shape and aid in intracellular transport.
C) Lamin is found in the nucleolus, playing a crucial role in ribosome synthesis and processing.
D) Lamin is localized at the plasma membrane of the cell, contributing to cell adhesion and communication with the extracellular matrix.
b. SEM
The image to the right of grain structure in a metal was most likely collected with which type of microscope:
a. Confocal
b. SEM
c. TEM
d. Brightfield
e. Fluorescence

b. SEM
The image to the right of electro spun polymeric fibers was most likely collected with which type of microscope:
a. Confocal
b. SEM
c. TEM
d. Brightfield
e. Fluorescence

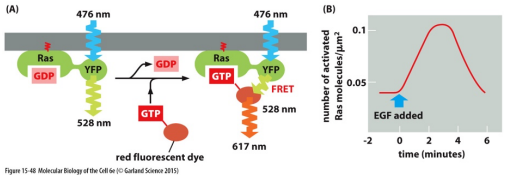
e. All of the statements are correct.
Which statement below is false (not correct) of the data in the figure shown above:
a. The approach being used is based on fluorescence resonance energy transfer (FRET).
b. When spaced very close together, the emission light from the YFP domain of Ras provides the excitation light for the red fluorescent dye conjugated to the GTP molecule.
c. EGF is a growth factor that binds cell surface receptors and activates Ras molecules.
d. The data shows that GTP binding to Ras occurs within 6 minutes following the addition of EGF.
e. All of the statements are correct.
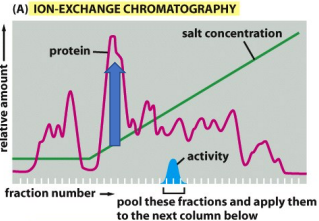
c. The protein identified at this point interacts weakly with the porous solid matrix of the column.
The graph shown at right suggests which of the following about the protein content at the point indicated by the blue arrow:
a. The protein content in the sample at this point (identified by the blue arrow) interacts strongly with the porous solid matrix of the column.
b. The protein identified at this point does not interact with the porous solid matrix of the column.
c. The protein identified at this point interacts weakly with the porous solid matrix of the column.
d. The protein identified at this point contains the enzyme being purified.
a. Confocal
Lea, a biomedical engineering student, is investigating the effects of a new drug on tissue scaffolds designed for regenerative medicine. Specifically, she is studying how the drug affects the alignment and growth of fibroblast cells within the scaffold, which is crucial for improving tissue regeneration. The scaffold is embedded with fluorescent markers that bind to specific cellular components like the cytoskeleton and extracellular matrix. Select the best microscope that will allow Lea to assess how the fibroblast cells are organized within the scaffold and track their structural changes over time, in three-dimensional images of the cells at various depths.
a. Confocal
b. SEM
c. TEM
d. Brightfield
e. Fluorescence
c. TEM
Roger, a biomedical engineering graduate student, is conducting research on the cellular mechanisms of Alzheimer's disease. Specifically, he is investigating how amyloid-beta plaques accumulate within the neurons and how they disrupt cellular processes. In order to understand the relationship between amyloid-beta and key cellular organelles, Roger needs to visualize the protein aggregates at the ultrastructural level within the neurons.
a. Confocal
b. SEM
c. TEM
d. Brightfield
e. Fluorescence
d. Brightfield
Julia, a biomedical engineering student, is researching the effects of a new therapeutic compound on human epithelial cells. Her project focuses on understanding how this compound influences cell proliferation and morphology. Specifically, she is studying cultured human keratinocytes, which are important for wound healing and skin regeneration. To observe the effects of the compound, Julia prepares a series of cell cultures, each treated with different concentrations of the drug. She needs to monitor the growth, shape, and general behavior of the keratinocytes over time to determine if the compound influences cell division or alters cellular structures.
a. Confocal
b. SEM
c. TEM
d. Brightfield
e. Fluorescence
B) Affinity Chromatography
Which of the following techniques involves the use of a specific ligand covalently linked to an insoluble matrix to selectively bind and purify proteins from a complex mixture?
A) Mass Spectrometry
B) Affinity Chromatography
C) X-ray Crystallography
D) SDS-PAGE
B) 2D NMR allows the distinction of overlapping peaks in larger molecules
What is the primary advantage of using a 2D NMR spectroscopy technique over a 1D NMR spectroscopy technique?
A) 2D NMR provides higher resolution for small molecules
B) 2D NMR allows the distinction of overlapping peaks in larger molecules
C) 2D NMR requires smaller sample volumes
D) 2D NMR is less dependent on functional groups
D) MS/MS allows for the determination of the amino acid sequence of peptides by fragmenting the peptide and analyzing the resulting fragments using two mass analyzers, which provides detailed structural information.
Which of the following statements accurately describes the use of mass spectrometry (MS) and tandem mass spectrometry (MS/MS) in protein analysis?
A) Mass spectrometry (MS) is used primarily for protein sequencing, while tandem mass spectrometry (MS/MS) is mainly used for quantification of known proteins.
B) In MS, the ion source generates peptide ions that are directly exposed to high-energy gas for fragmentation, while MS/MS involves the use of a single mass analyzer to separate ions.
C) The primary function of MS is to identify post-translational modifications in proteins, while MS/MS is used solely for simple identification of purified proteins.
D) MS/MS allows for the determination of the amino acid sequence of peptides by fragmenting the peptide and analyzing the resulting fragments using two mass analyzers, which provides detailed structural information.
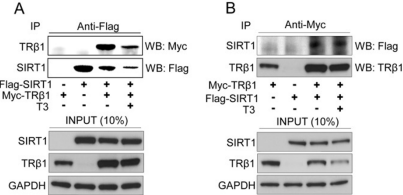
False
T/F. According to the data shown to the right, GAPDH binds SIRT1.
a. Yes
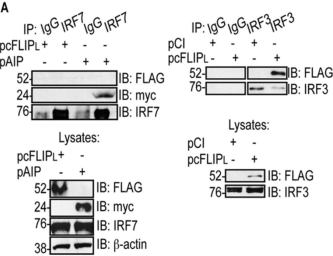
In the data shown to the right, plasmids were used to transfect cells with a FLAG-tagged cFLIPL (pcFLIPL), a myc-tagged AIP protein (pAIP), and an HA-tagged CL.
IRF7 and IRF3 are naturally expressed by these cells.
a. Yes
b. No
c. Can’t be determined
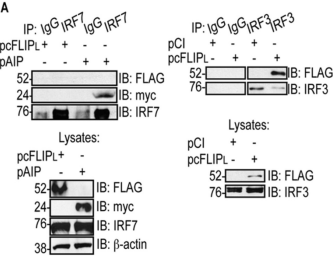
c. Can’t be determined
In the data shown to the right, plasmids were used to transfect cells with a FLAG-tagged cFLIPL (pcFLIPL), a myc-tagged AIP protein (pAIP), and an HA-tagged CL.
T/F. Using the same data above, does CL bind to IRF3?
a. Yes
b. No
c. Can’t be determined
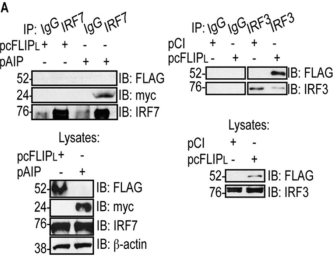
a. Yes
In the data shown to the right, plasmids were used to transfect cells with a FLAG-tagged cFLIPL (pcFLIPL), a myc-tagged AIP protein (pAIP), and an HA-tagged CL.
T/F. Using the same data above, does cFLIPL bind to IRF3?
a. Yes
b. No
c. Can’t be determined
False
T/F. According to the data shown to the right, Gab1 expression is greater than that of either SHP2 or p85 PI 3-K.
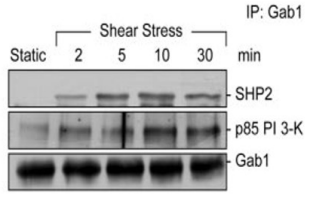
False
T/F. According to the same data shown above, SHP2 and p85 PI 3-K bind to Gab1 in sequence (meaning one after the other).
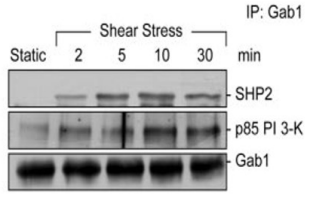
False
T/F. The data below shows that TbTim17 and Trap1 bind strongly together. Ft refers to the sample fraction that flows through the beads without binding to the Ab. B refers to the bound fraction.
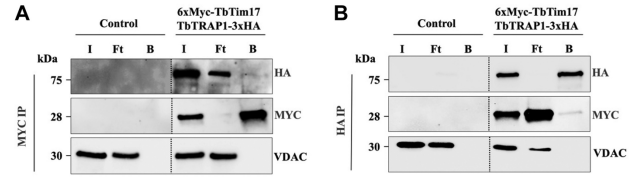
False
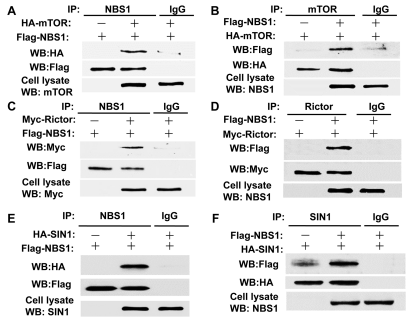
T/F. The data below, in panel A, shows that NBS1 binds to HA.
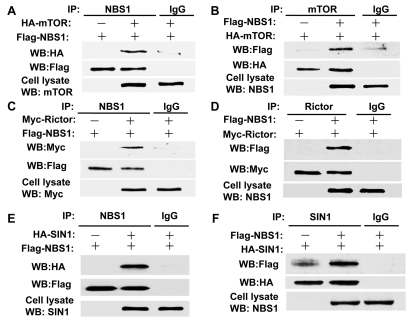
True
T/F. According to the same data shown above, it is reasonable to assume that SIN1 forms a complex with mTOR.
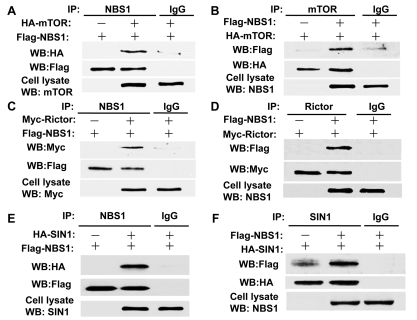
False
T/F. According to the same data shown above, in panel A, IgG prevents the binding between mTOR and NBS1.
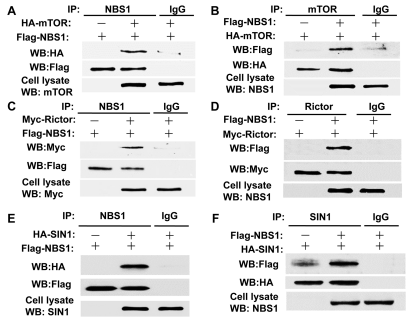
False
T/F. According to the same data shown above, in panel E, HA binds to FLAG