Section 3.1: Transporters
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
The Druggable Proteome
25% of the human genome codes for membrane proteins (eg. channels, transporters, ion pumps, etc.)
67% of known drug targets are membrane proteins
this is b/c they control essential physiological processes and are accessible to drugs (on outside of cells, drug doesn’t need to enter the cell first)
binding sites are often well-defined and modulating them produces large, fast effects
The Druggable Proteome: Examples of Membrane Proteins as Drug Targets
stomach ulcers: drugs that target the gastric H+/K+ ATPase (proton pump in the stomach lining that acidifies the stomach) → reduces stomach acidity
Type II diabetes: drugs targeting the K+ channel in β pancreatic cells → higher insulin secretion
Hypertension: drugs acting on water channels in the kidney → reduce salts reabsorption
Stress: drugs that target the β-adrenoreceptor (β-blockers) → less production of adrenaline
Permeability Properties of Synthetic Lipid Bilayer
molecules can diffuse across the lipid bilayer down its concentration gradient
size: smaller molecules diffuse more easily
polarity: the bilayer’s interior is hydrophobic (oil-like), so nonpolar/hydrophobic molecules cross easily and rapidly
gases (O2, CO2) and small uncharged molecules (urea, ethanol) can move by diffusion
charged molecules (ions and larger) are very impermeable, no matter how small (eg. Na+, K+, Cl-)
hydration shells prevent them from entering the hydrophobic membrane
Passive Transporters Part 1
move molecules down their concentration gradient without using energy
direction is determined by the concentration gradient (always high → low)
there's a finite number of transporters, so increasing substrate beyond a certain point doesn’t increase rate indefinitely (saturable)
binding is typically 1:1, one substrate at a time
transport is specific, each transporter recognizes particular substrates
no chemical bonds are made or broken in the conversion
Passive Transporters Part 2
affinity for substrate is the same on both sides; the transporter can work in either direction (process is fully reversible)
cannot concentrate substrate inside, only moves it to balance the gradient
passive transporters are for large hydrophilic substrates (eg. sugars, amino acids, vitamins)
these molecules can’t diffuse, so they require transporters
rate of transport is faster than diffusion
rate depends on concentration gradient (more substrate outside = faster), and conformational change speed (flip of transporter between outward and inward facing states)
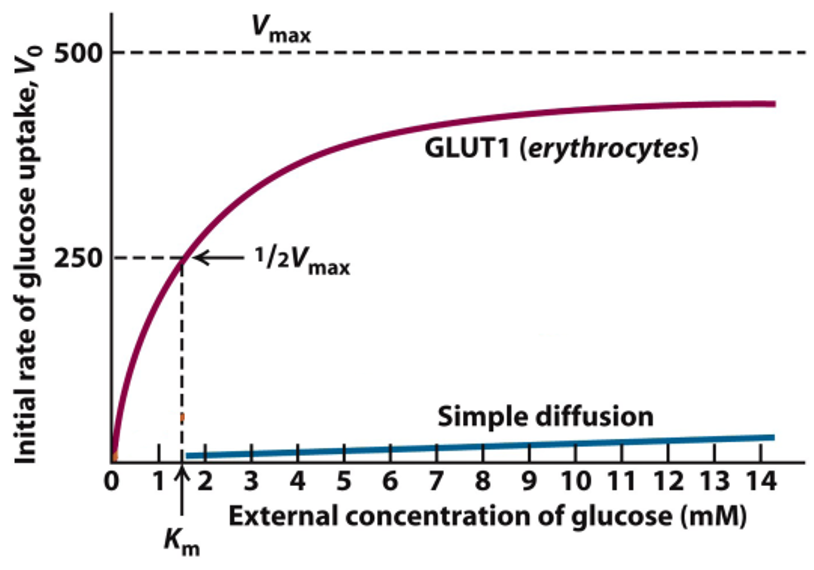
Kinetics of Glucose Transport in RBCs: GLUT1
different Km → different transports behaviors (Km reflects affinity)
RBCs need a constant glucose uptake
GLUT1 Km = 1.5 mM, blood glucose is 5mM
at 5mM, there is import of glucose close to Vmax
below 5mM, GLUT1 in RBCs still imports glucose efficiently
above 5mM, transport of glucose is limited
after import, glucose is phosphorylated to glucose-6-phosphate, which has no affinity for GLUT1
this traps glucose inside, and makes import one-way/irreversible
accounts for 2% of the protein in the PM of RBCs
Km
substrate concentration at which an enzyme-catalyzed reaction reaches half its maximum velocity (Vmax)
measure of how efficiently an enzyme works
low Km = high affinity → transporter works well even at low substrate conc.
high Km = low affinity → transporter mainly active when substrate conc. is high
Kinetics of Glucose Transport in RBCs: GLUT2
high Km, only active when glucose is high (eg. after a meal)
liver stores it as glycogen
the more glucose, the more transport into the liver
transports glucose out of the hepatocytes (glucose storage) to replenish blood glucose
import of glucose is quasi-linear (curve looks like simple diffusion)
at low glucose (below 5mM), glucose is released from hepatocytes, and GLUT2 does not import
has high Km (~15mM) → low affinity
in the liver and β-cells of pancreas
Kinetics of Glucose Transport in RBCs: Figure
Enzyme Kinetics
even though transporters are not enzymes, their behavior follow Michaelis-Menten-like kinetics because:
the transporter binds a substrate, it undergoes a conformational change, and it can become saturated
Vmax: the maximum rate of transport, when all transporters are fully occupied with substrate
reflects how fast the transporter flips between outward-open and inward open states
Km: substrate conc. where the rate is ½ Vmax
Enzyme Kinetics: Measuring Kinetics of Glucose Transport
measure the initial rate of glucose uptake (V0)
V0 depends on external glucose concentration
at high glucose, transport reaches Vmax; all transporters are occupied → saturation
when the rate of uptake is ½ Vmax, the transport if half-saturated
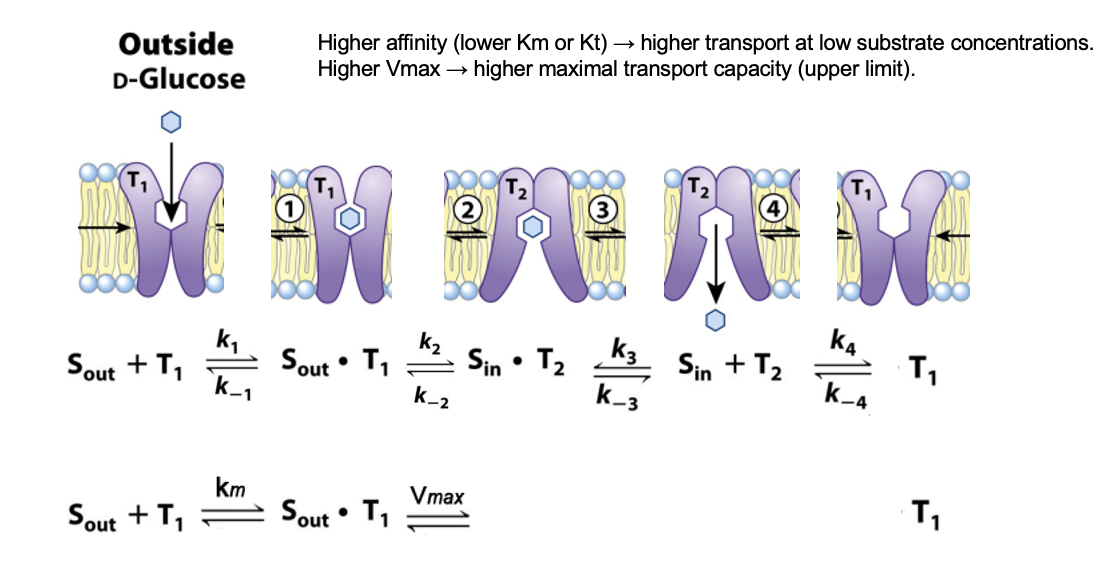
GLUT: Conformations
the transporter exists in two conformations
T1: the glucose binding site exposed on the outer surface of the PM
T2: the binding site exposed on the inner surface
Glucose Transport Steps
glucose binds to T1
binding lowers the activation energy for T1 → T2 transition
the transporter flips its conformation
in T2, glucose is released into the cytoplasm
release allows the transporter to return to the T1 conformation
The Isoforms of the Glucose Transporter
the human genome encodes at least 12 highly homologous GLUT proteins
each transporter has a unique distribution
the same function, but different affinity for glucose, gives different transport kinetics
GLUT3 is expressed in neuronal cells, which depend on a constant influx of glucose (5-fold higher affinity than GLUT1)
GLUT4 in skeletal (Km ~5 mM): presence in the membrane is regulated by insulin
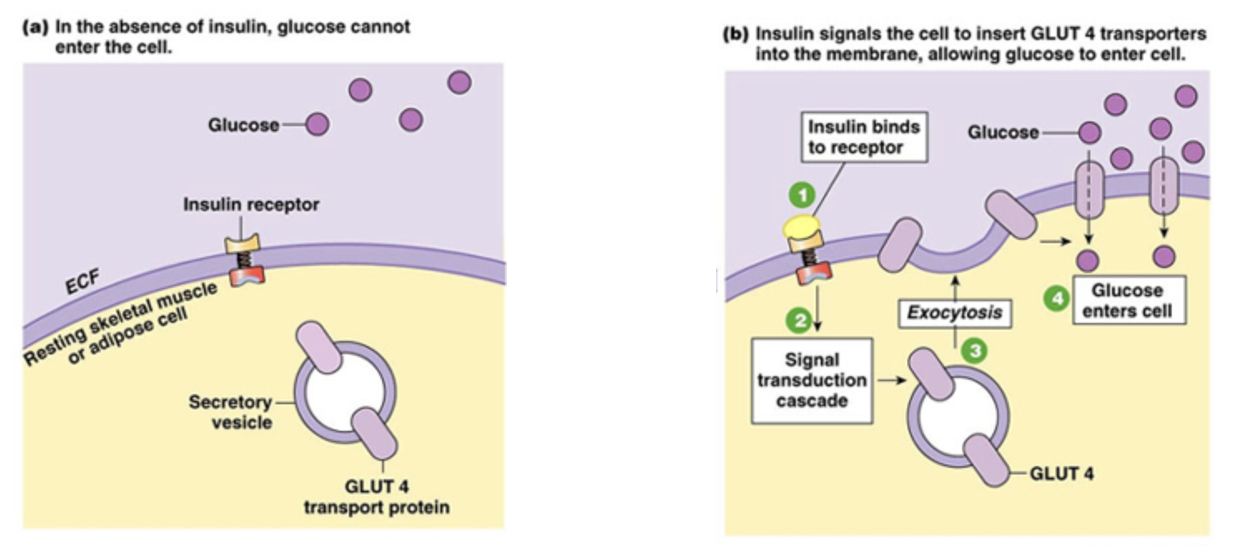
Increase of Glucose Transport with Insulin: What Normally Happens
Between meals: GLUT4 transporters are stored in intracellular vesicles in muscle cells and adipocytes
very little glucose uptake at the cell surface
After a carbohydrate rich-meal: blood glucose rises above ~5mM
pancreas releases insulin, triggering GLUT4-containing vesicles to move to and fuse with the PM
more GLTU4 on surface → 15x increase in glucose uptake
excess glucose is stored as glycogen in muscle or TAG in adipocytes
when insulin levels drop, GLUT4 is endocytosed back into vesicles
Increase of Glucose Transport with Insulin: Type 1 Diabetes
in type 1 (insulin-dependent) diabetes mellitus, insulin is not released and GLUT4 remains in vesicles
autoimmune destruction of β-cells → no insulin production
cells do not take up glucose → blood sugar stays high
insulin injections restore the signal → GLUT4 moves to the surface
Increase of Glucose Transport with Insulin: Type 2
insulin is produced, often in high levels
but cells do not respond → signaling pathway fails
GLUT4 does not efficiently move to the surface
insulin injections are less effective because the signaling pathway is the issue, not the insulin level
Increase of Glucose Transport with Insulin FIGURE
SGLT Transporter
Sodium Glucose Transporter (Symporter)
uses energy stored in sodium gradient to import glucose against its concentration gradient
outside the cell: High Na+, inside the cell: low Na+
since Na+ strongly wants to enter the cell (downhill energetically), SGLT couples this downhill movement of Na+ to the uphill transport of glucose
this means SGLT can import glucose even when extracellular glucose is low
Na+ gradient was created by the Na+/K+ ATPase
binding of Na+ and glucose is cooperative. If Na+ isn’t bound first to increase the glucose affinity of the transporter, glucose won’t bind well
SGLT Transporter: Stoichiometry
2 Na+ ions + 1 glucose transported inward together
the binding of Na+ increases the affinity for glucose → ensures glucose gets captured even at low concentrations
SGLT Transporter: Transport Cycle
1) Outward facing conformation (open to outside)
binding sites for Na+ and glucose are exposed to exterior
2) Occluded conformation: both gates are closed, no access to either side
transporter flips to face inside
3) Inward-facing conformation (open to inside)
binding sites now face the cytoplasm
4) Once transporter is empty, the transporter resets back to outward-facing
SGLT Transporter: Transport Cycle Figure

Energy Stored in an Ion Gradient
for ions: ∆G = ∆Gm + ∆Gc
∆G is how much energy is available when Na+ moves into the cell
∆Gc = concentration gradient contribution, this is the energy due to the difference in concentration of Na+ across the membrane
∆Gm = electrical gradient concentration, this is the energy due to the membrane potential
inside of the cell is typically negative relative to the outside
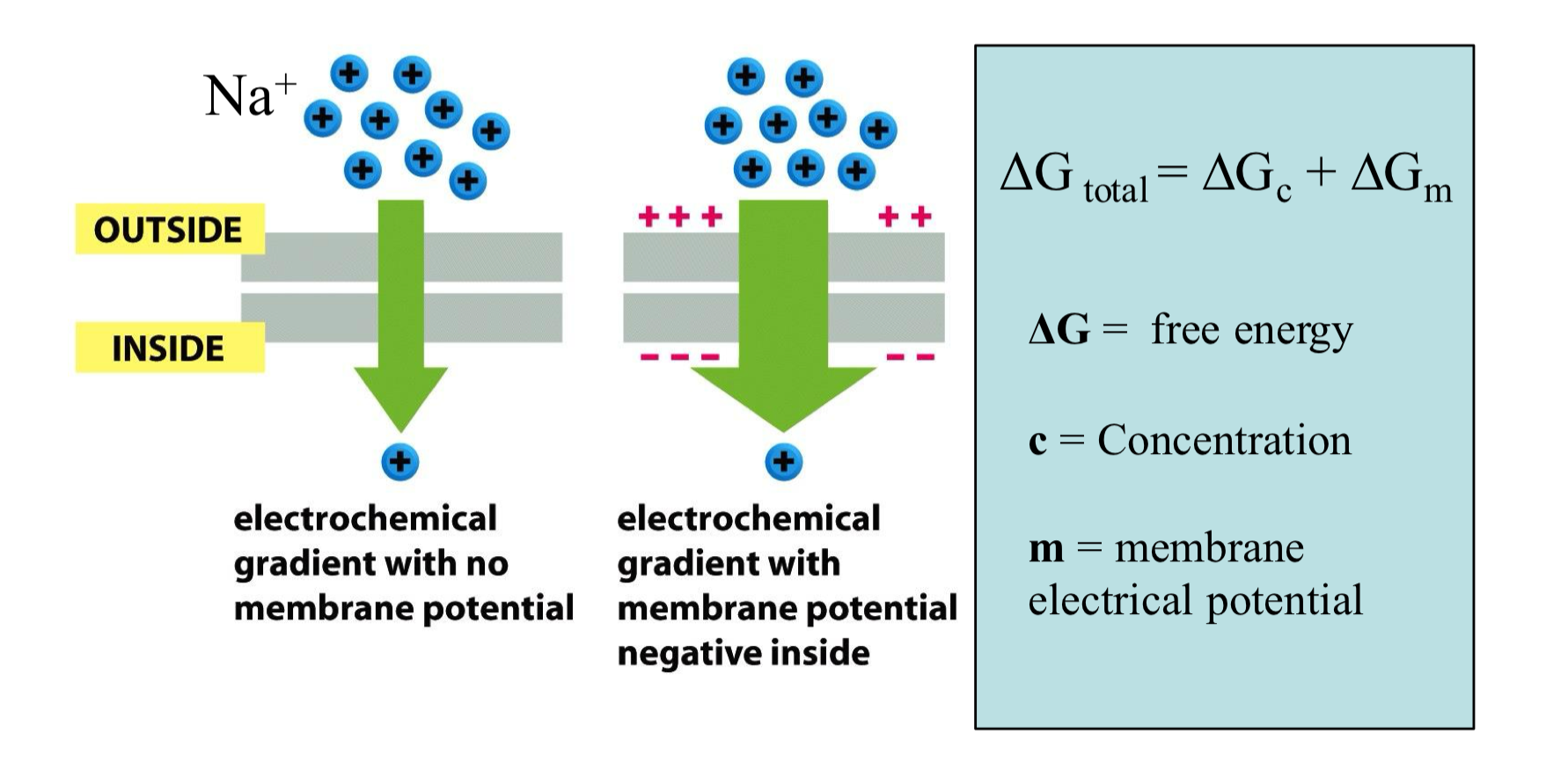
Energy Derived from Na+ Gradient
the movement of Na+ across the membrane is driven by the concentration gradient and the membrane electric potential
both point forces inward, so the total free energy change for Na+ is highly favorable
∆Gc = RT ln( [Nain] / [Naout]
∆Gm = FE
E: membrane electric potential = -70mV
F: Faraday constant: 23062 cal/molV
![<ul><li><p>the movement of Na<sup>+</sup> across the membrane is driven by the concentration gradient and the membrane electric potential</p></li><li><p>both point forces inward, so the total free energy change for Na<sup>+</sup> is highly favorable</p></li><li><p>∆G<sub>c</sub> = RT ln( [Na<sub>in</sub>] / [Na<sub>out</sub>]</p></li><li><p>∆G<sub>m</sub> = FE</p><ul><li><p>E: membrane electric potential = -70mV</p></li><li><p>F: Faraday constant: 23062 cal/molV</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/2af762aa-822b-4337-982d-9f84c61ff214.png)
LacY Transporter
LacY is a proton-driven symporter in bacteria , where it uses an ion (H+ gradient) gradient as the energy source
417 amino acids long, but only 3 are essential
some residues serve for lactose binding, some for proton transport
moves a sugar (lactose) against its concentration gradient
has 12 TM helices forming a central cavity in the middle of the membrane (where lactose binds)
switches between outward facing and inward facing states
the 6 N-ter and 6 C-ter produce a structure with a rough two-fold symmetry
requires cooperative binding of ion and sugar
in the cytosol, β-galactosidase cleaves lactose into glucose and galactose
LacY Figure
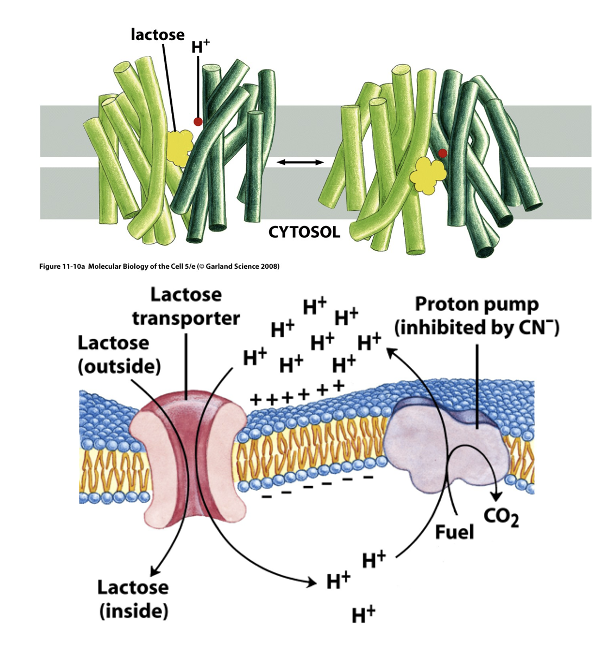
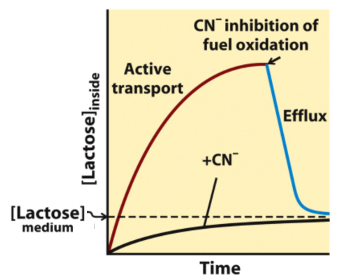
LacY Inhibition: Cyanide
when cyanide is added in the middle of the experiment, lactose goes back out
blocks transport of lactose
cyanide blocks ETC which makes the proton gradient
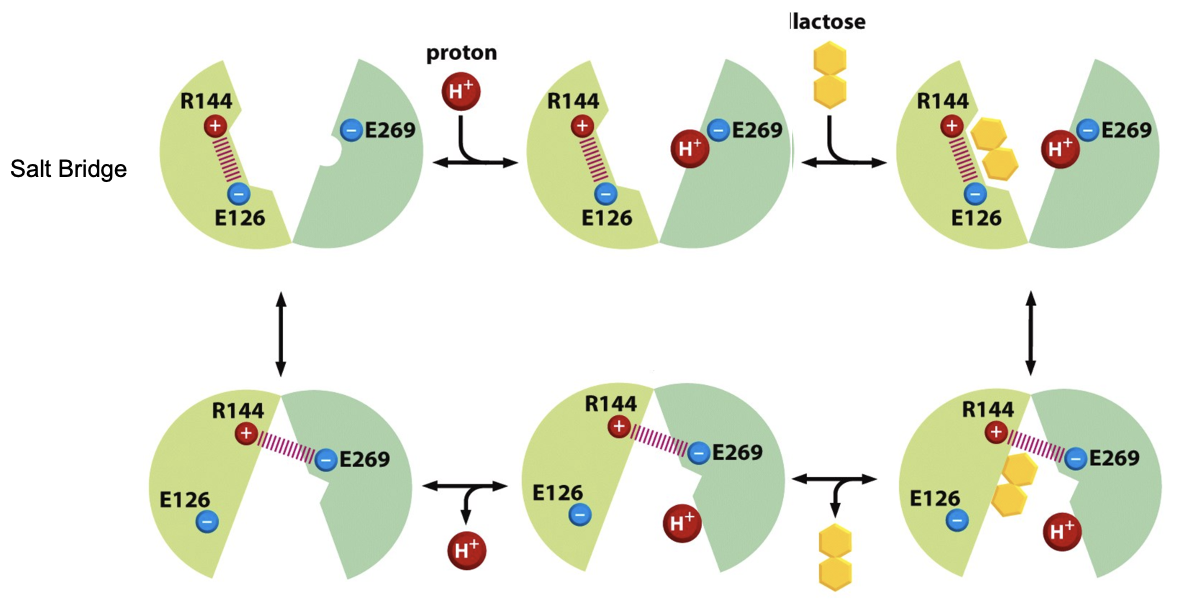
The PMF drives Transport of Lactose
essential a.a’s: E269 (Glu), R144 (Arg)
H+ loading is favored b/c E269 attracts H+
H+ binding increases lactose affinity
the presence of both substrates favors the electrostatic interaction of R144 with E269
the electrostatic bond triggers conformational change, tilting the transporter from the outward facing → inward facing
inside the cell, the cytosol is more basic and E269 deprotonates
the loss of H+ weakens lactose binding → lactose is released
once H+ is released, the salt bridge breaks and the transporter resets to outward facing
the process is fully reversible, lactose can escape the cell
The PMF drives Transport of Lactose FIGURE
Antiporter The Cl- / HCO3- (AE1): Background
enables CO2 transport between tissues and lungs
called anion exchanger 1 and contributes up to 25% of RBC membrane proteins
HCO₃⁻ OUT ↔ Cl⁻ IN (in tissues)
HCO₃⁻ IN ↔ Cl⁻ OUT (in lungs)
Antiporter The Cl- / HCO3- (AE1): Step 1
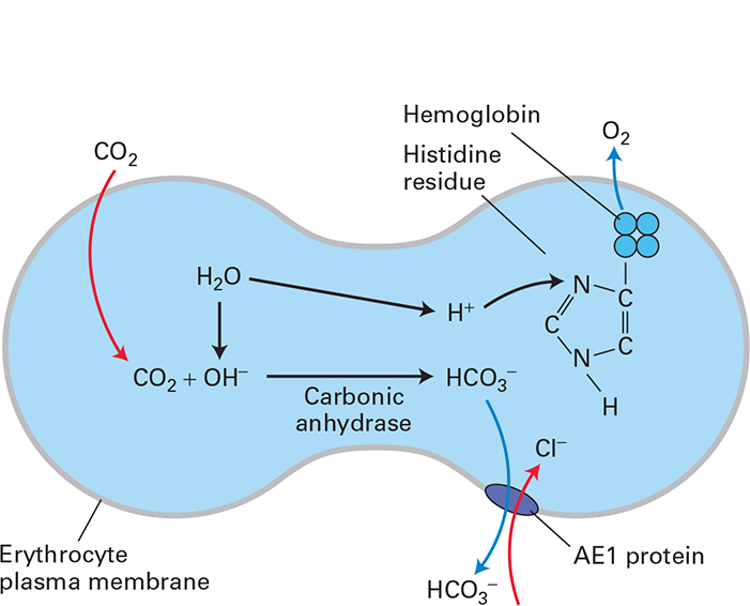
In respiratory tissues (where CO2 is produced)
waste CO2 diffuses into RBCs and is converted to HCO3- by carbonic anhydrase
HCO3- must leave the cell (prevent build up), otherwise reaction stops. It is more soluble in blood plasma than CO2 (better carried to the lung)
AE1 exports it and imports Cl- (1:1 exchange)
this massively increases CO2 transport efficiency
Antiporter The Cl- / HCO3- (AE1): Respiratory Tissue Figure
Antiporter The Cl- / HCO3- (AE1): Step 2
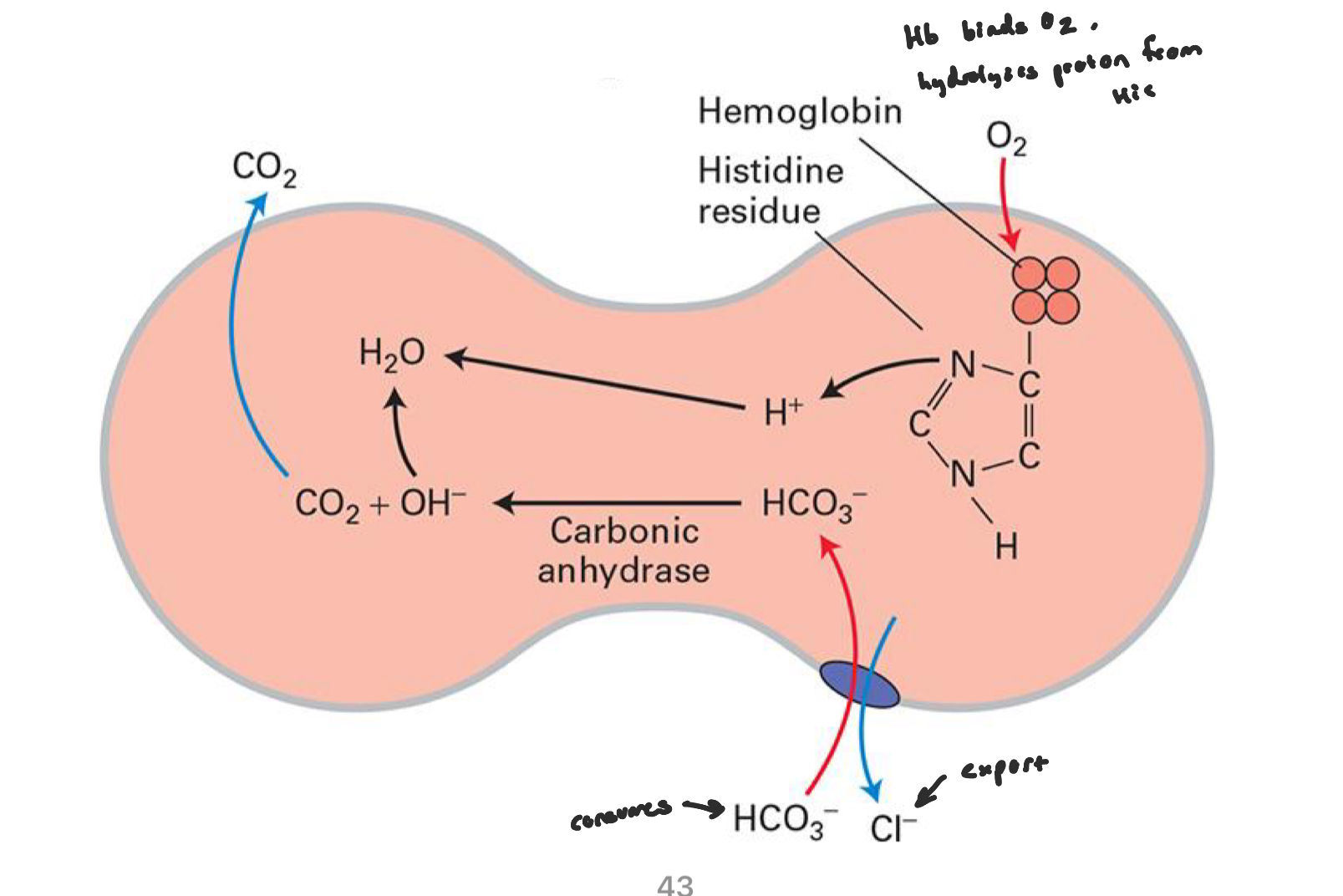
In lungs (where CO2 is exhaled)
HCO3- reenters RBC through AE1 and Cl- leaves
carbonic anhydrase converts HCO3- → CO2 → exhaled
Antiporter The Cl- / HCO3- (AE1): Lung Figure
Antiporter The Cl- / HCO3- (AE1): Membrane Potential
even though ions move, AE1 exchanges one negative ion for another
no net charged moved
membrane potential stays constant (electroneutral transport)
Antiporter The Cl- / HCO3- (AE1): Obligatory Coupling
AE1 cannot move just HCO3- or just Cl-
if Cl- is absent, HCO3- transport stops
Antiporter The Cl- / HCO3- (AE1): Hemoglobin
when Cl- enters the RBC in tissues, it slightly lowers the cystolic pH (more acidic) and Hb released O2 more readily at low pH
when Hb reaches the lungs and O2 binds, the structural changes makes certain His residues release H+
this H+ reacts with HCO3- to form CO2 to be exhaled
Symporter vs. Antiporter
both cotransporters in secondary active transport, but differ in direction
symporter: move both substrates in the same direction
antiporter: move substrates in opposite directions
Primary Active Transport
directly uses cellular energy, primarily from ATP, to move substances (e.g Na+, K+) across membranes against their concentration gradient
use transmembrane proteins (pumps) that change shape after binding ATP, thus pumping molecules out or in
eg. Na+/K+ ATPase
Facilitated Diffusion
Type of passive transport where molecules move across a cell membrane down their conc. gradient, with the help of a specific carrier or channel protein
doesn’t require ATP
eg. Aquaporins, GLUT1
ABC Transporter Family
ATP binding casette
48 types in humans; mostly exporters (e.g bile salts, lipids), many are linked to disease
in bacteria, 5% of the genome encodes for ABC transporters, including importers of nutrients
each transporter is specific for a class of substrates (e.g sugar, vitamins, a.a’s, proteins)
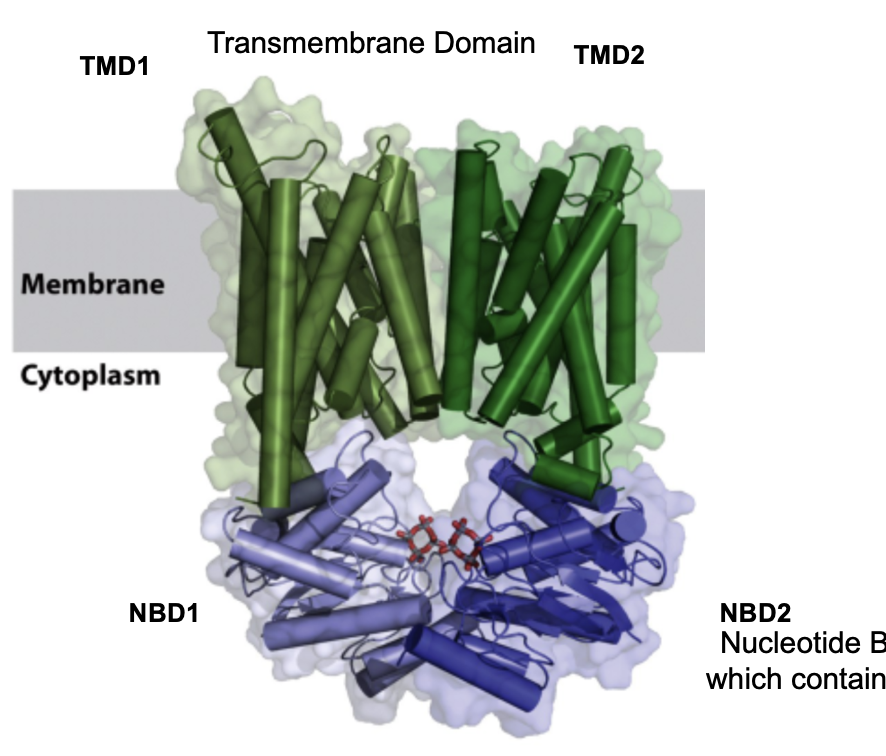
share a conserved structure: nucleotide binding domain (NBDs) for ATP binding/hydrolysis, transmembrane domains (TMDs) that form the transport pathway
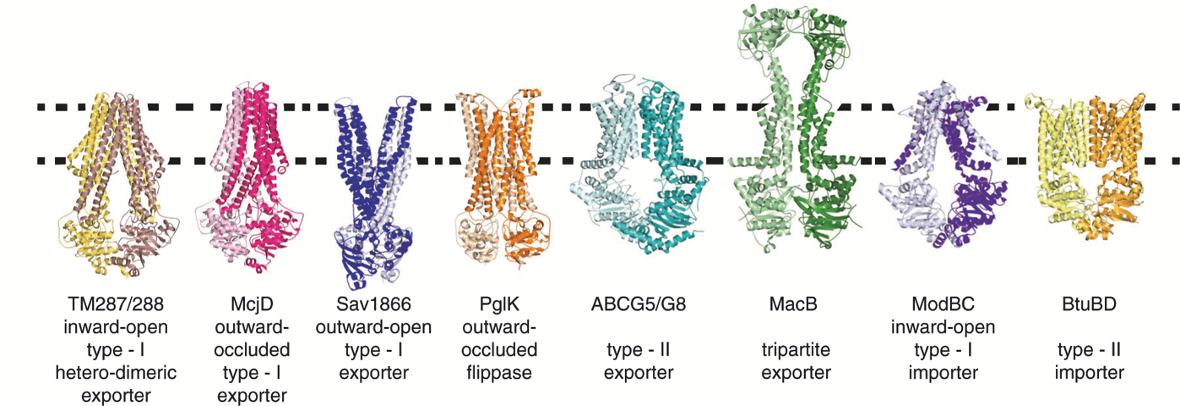
Structural variations found across ABC Transporters
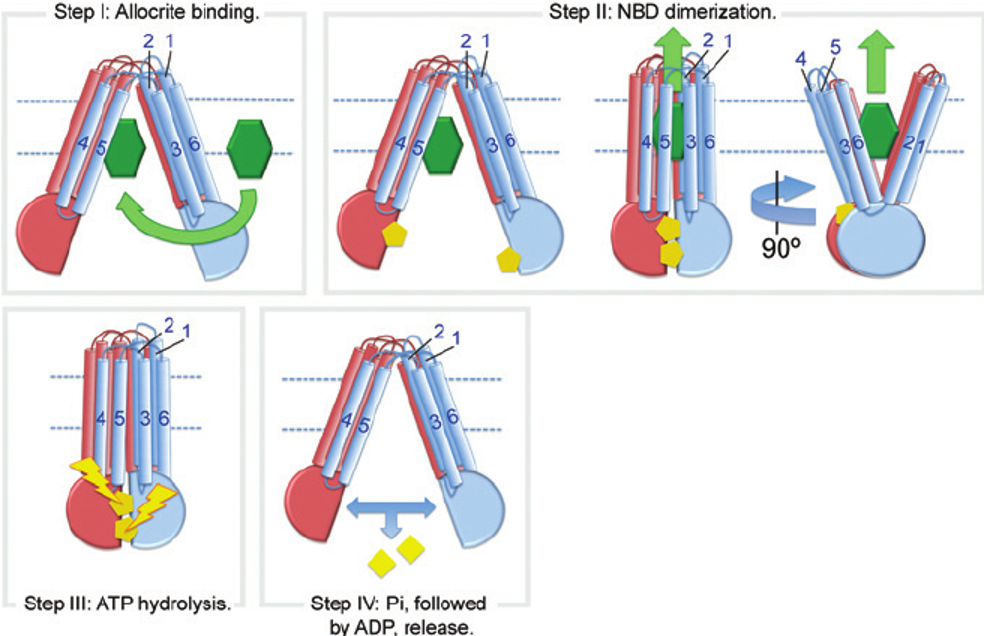
General Mechanism for ABC Transporters: Steps
substrate binding to TMDs on one side of membrane
NBD dimerization: 2 ATP molecules bind and are sandwiched at the NBD dimer interface, causing it to dimerize
ATP hydrolysis → Pi released first then ADP
Resetting the transporter: NBDs dissociate, then transporter returns to original conformation
General Mechanism for ABC Transporters: Steps FIGURE
ABCB1 (MDR1)
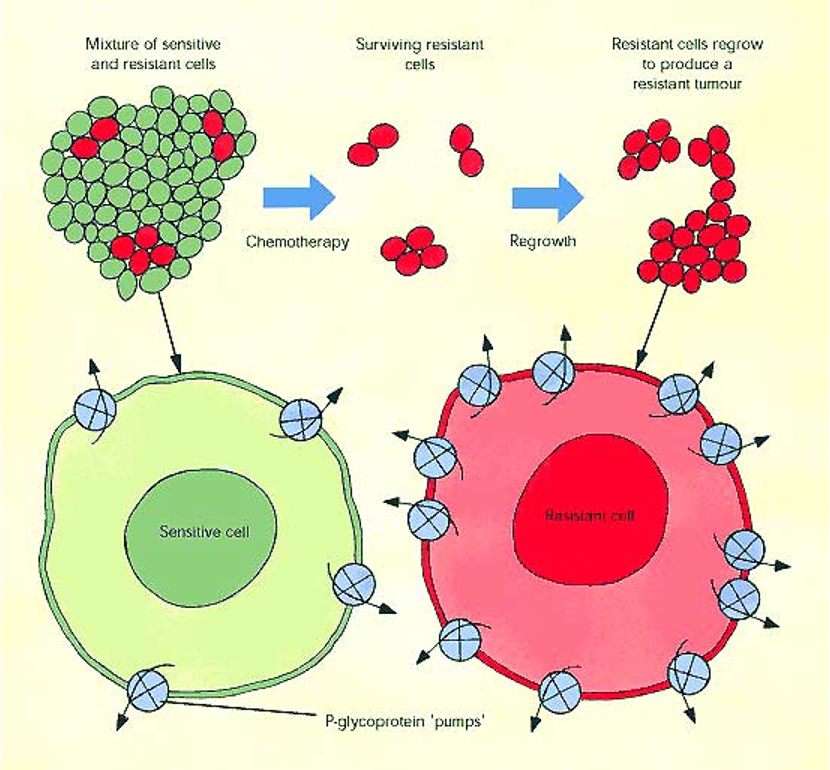
Multidrug Resistance Protein
ABCB1 is an ABC transporter that normally exports hydrophobic metabolites into bile or urine
in cancer: tumor cells overexpress ABCB1, leading to resistance to multiple chemotherapic drugs
chemotherapy selects for cells with high ABCB1 expression → these cells survive and overgrow
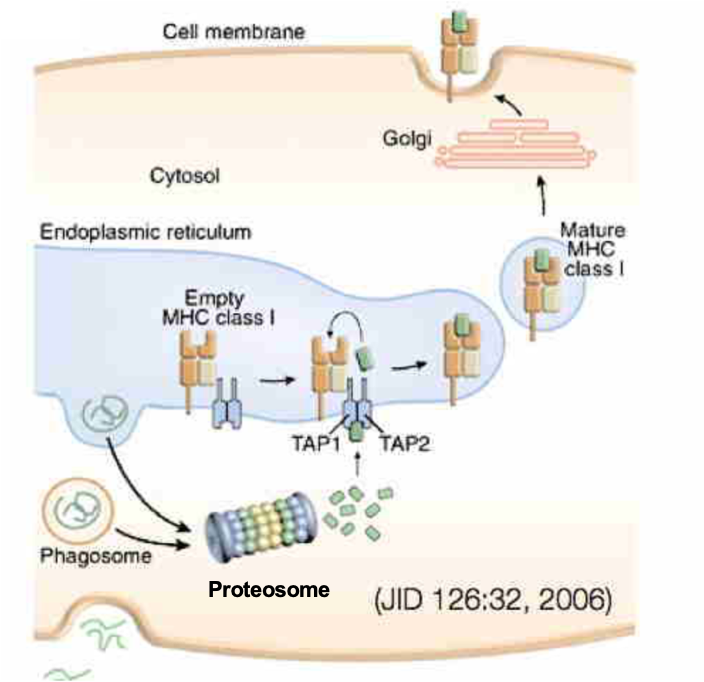
ABCB2 TAP Transporter
TAP is an ABC transporter composed of TAP1 and TAP2
plays central role in adaptive (not innate) immunity
foreign proteins enter the cell and are degraded in cytosol into short peptides
these peptides are recognized and transported by TAP1-TAP2 into ER lumen → peptides are loaded onto MHC class 1 molecules
peptides are not taken up by Sec61 (don’t have signal peptide)
Peptide-MHC 1 complexes traffic to the PM
CD8+ cytotoxic T cells recognize the foreign antigen and kill the cell (immune surveillance)
ABCB2 TAP Transporter FIGURE
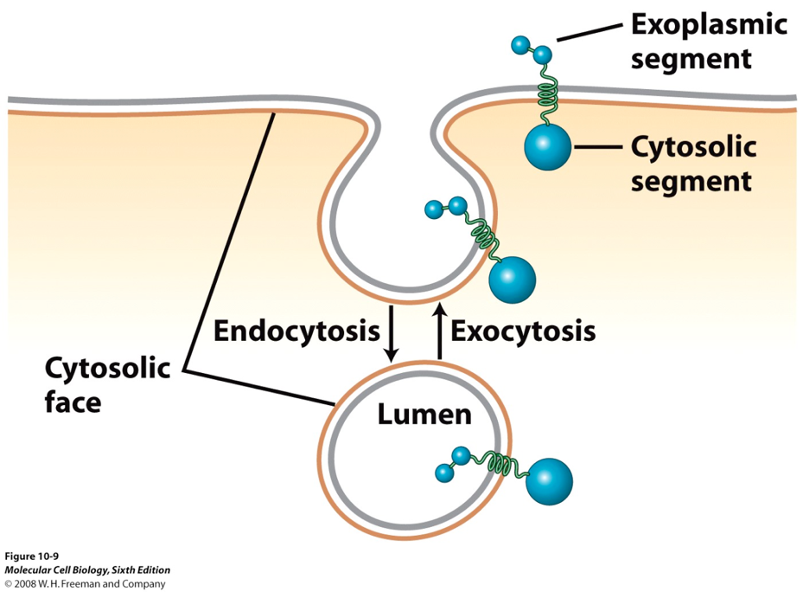
Orientation of MHC Molecules
MHC 1 proteins are synthesized in ER membrane w/ a fixed orientation
luminal side of ER = extracellular side after secretion, cytosolic domains always face the cytosol
during transport (ER → Golgi → PM), proteins move in vesicles
vesicle fusion does NOT flip membranes (cytosolic leaflet remains cytosolic, luminal leaflet becomes extracellular)
in other words, the fusion of a membrane vesicle to the cell surface maintains the membrane polarity
ABCC7-CFTR
CFTR is in the ABC transporter family, but functions an ion channel, not a pump
forms a Cl- channel in the PM of epithelial cells
ATP binding (not hydrolysis) regulates channel gating
ATP binding to NBD = channel open
ATP release/hydrolysis = channel closes
Cl- moves passively thru CFTR down its electrochemical gradient
CFTR activity regulates ion and water balance in epithelial surfaces (e.g lungs)
mutations in the CFTR protein cause cystic fibrosis
Overall Pathogenesis of Cystic Fibrosis: Normal Lung
CFTR is located on apical surface of airway epithelial cells, allowing Cl- secretion into the airway lumen
Cl- movement draws water osmotically, maintaining a hydrated mucus layer
proper hydration allows cilia function and efficient clearance of particles
Overall Pathogenesis of Cystic Fibrosis: Cystic Fibrosis
defective or absent CFTR → reduces Cl- secretion
altered ion balance disrupts osmotic water movement
mucus becomes dehydrated and viscous
thick mucus traps bacteria → persistent infection and inflammation
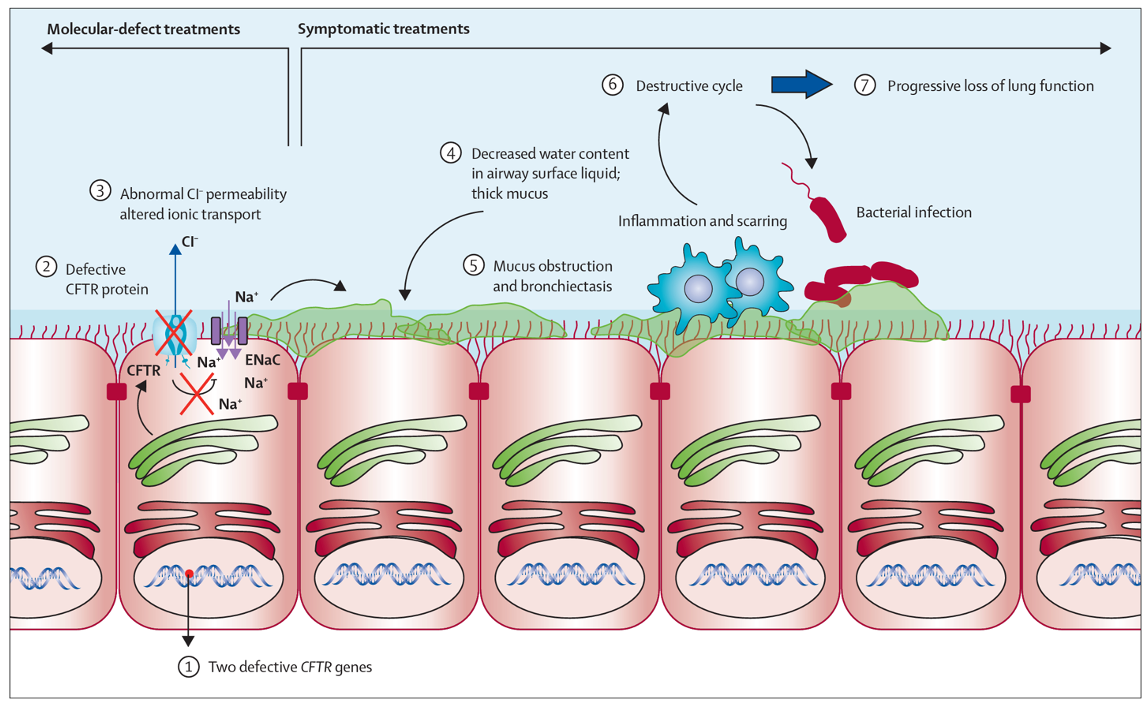
Over 300 mutations cause CF but ∆F508 in about 90% of patients
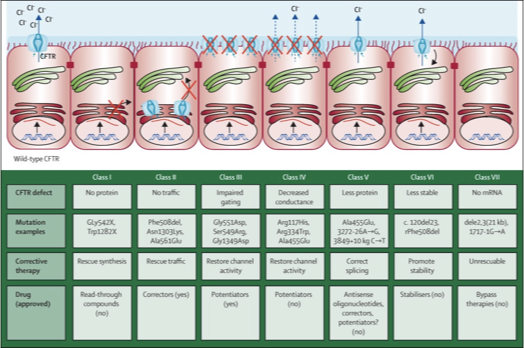
Precision Medicine
precision medicine treats disease by targeting the molecular defect, not just the symptoms
Trikafta is a combination of ivacaftor + tezcaftor + elexcaftor
these drugs act as CFTR correctors
tezcaftor + elexcaftor → improve CFTR folding and trafficking to the membrane
Ivacaftor → increases channel opening (gating) of CFTR already at the membrane
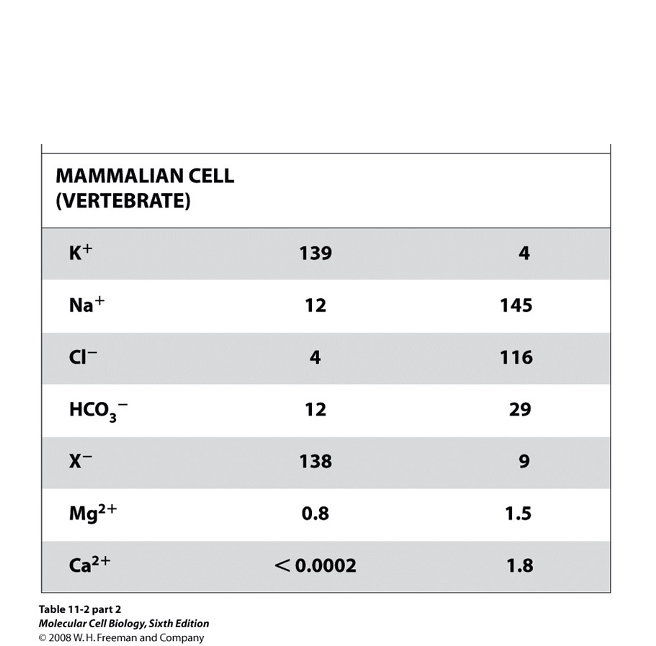
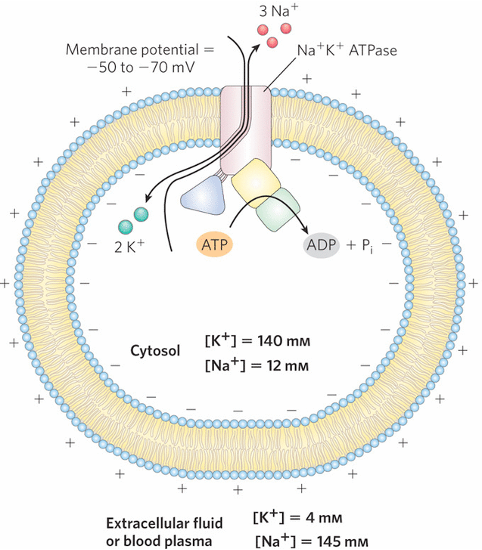
Typical Ion Concentrations in Mammalian Cells: Chemical & Electrical Gradients
Na+/K+ ATPase (P-type ATPase) establishes and maintains ion gradients across the PM
chemical gradient: created by unequal ion conc. (high K+ inside, high Na+ outside)
maintained by continuous ATP hydrolysis by the Na+/K+ pump
electrical gradient: the pump is electrogenic (net +1 charge leaves the cell each cycle) → inside of cell is electrically negative relative to ouside
additionally, the cytosol contains many (-) charged proteins
How Na+/K+ ATPase Works
maintains Na+ and K+ gradients across the PM
binds 3 Na+ from cytosol → ATP hydrolysis → conformation change → Na+ is expelled to extracellular space → 2 K+ ions bind from outside → dephosphorylation → conformation change → K+ released into cytosol
high K+ inside, high Na+ outside
contributes to resting membrane potential
the pump costs a lot of energy: constantly counteracting Na+ and K+ leakage
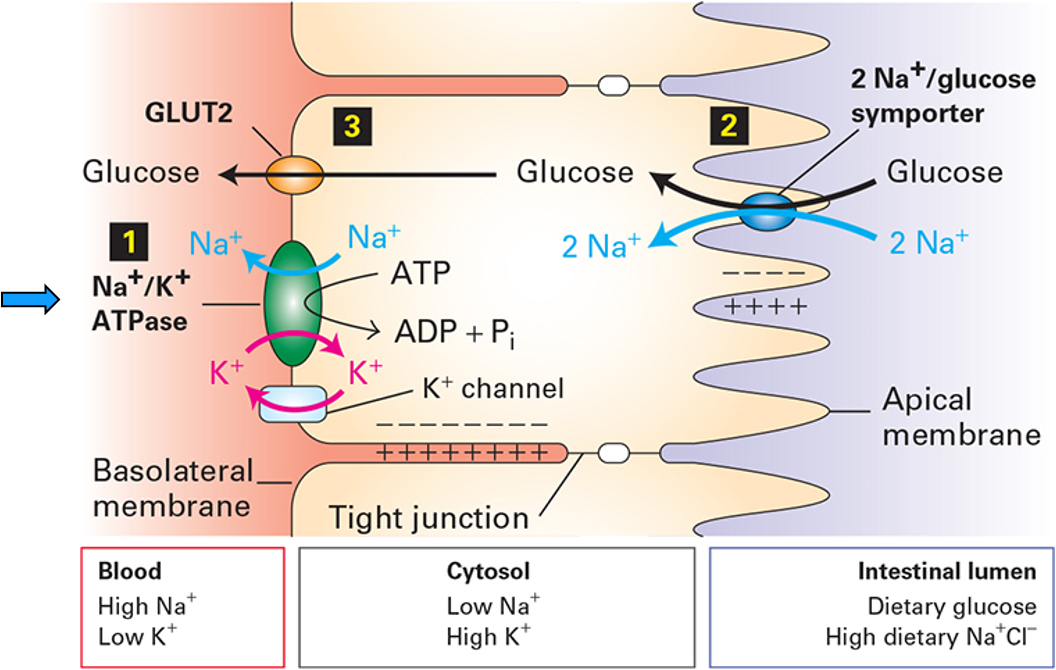
The Na⁺/K⁺ ATPase Drives Nutrient Transport
ATP is not used directly to transport glucose, it is used to create a Na+ gradient which then powers glucose uptake
strong inward Na+ electrochemical gradient that is the energy source for nutrient uptake
SGLT uses the Na+ gradient to import glucose (glucose is carried up its concentration gradient)
GLUT2 passively releases glucose into the bloodstream (down glucose gradient)
prevents glucose buildup
excess Na+ is removed (otherwise dissipates gradient) by Na+/K+ ATPase
The Na⁺/K⁺ ATPase Drives Nutrient Transport FIGURE
The Na+/K+ ATPase is Electrogenic FIGURE
P-type ATPase
P-type ATPases are phosphorylated by ATP
are cation pumps; maintain ionic differences b/w cytosol and extracytosolic environment
Na+/K+ ATPase is an antiporter for Na+ and K+
H+/K+ is an antiporter for H+ and K+ (acidifying stomach)
Ca2+ ATPase is an uniporter for Ca2+ (Ca2+ leads to muscular fiber contraction in myocytes).
P-type ATPase: SERCA (P-type ATPase)
sarcoplasmic ER calcium ATPase; located in sarcoplasmic reticulum (SR) of muscle cells
pumps Ca2+ from cytosol → SR lumen, using ATP (maintains low cytosolic Ca2+)
Muscle contraction cycle:
small Ca2+ influx across PM opens Ryanodine receptors (Ca2+ release channels) → Ca2+ binds to troponin → muscle contraction
relaxation: SERCA pumps Ca2+ back into SR, restoring low cytosolic Ca2+
V-type ATPase (V for vesicle/vacuolar)
pumps H+ into organelles (e.g. lysosomes, endosomes, vacuoles) and synaptic vesicles to acidify them
allows for lysosomal enzyme activity, vesicle trafficking, receptor-mediated endocytosis
1-2 V-type ATPases per vesicle
ATP-driven electrogenic pump: uses ATP hydrolysis to move H+ against their gradient
contains multi-subunit rotary: V1 domain (ATP hydrolysis), V0 domain (proton translocation)
H+ gradient drives neurotransmitter uptake via H+/neurotransmitter antiporter
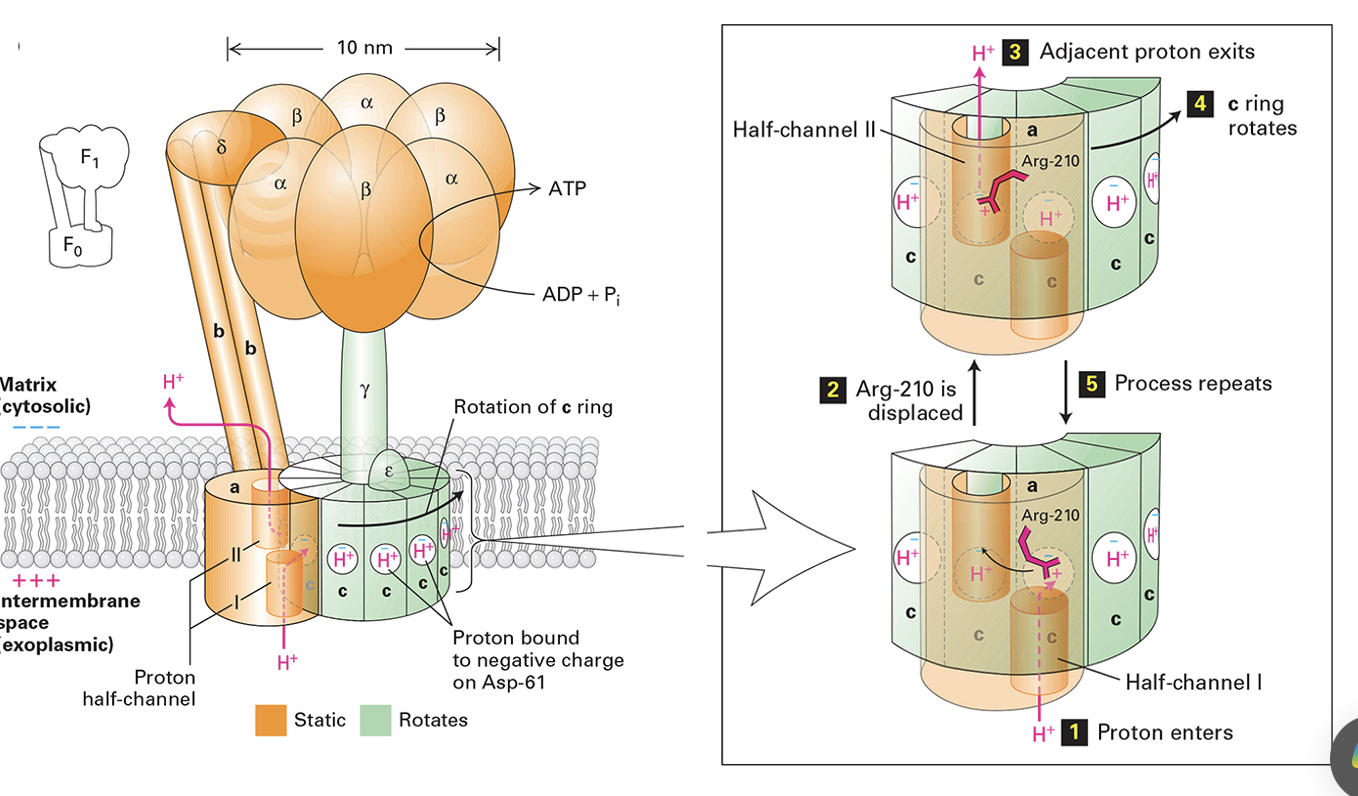
F-type ATPase
found in mitochondria, chloroplasts and bacterial PMs
primarily synthesizes ATP using the proton gradient (protons flow down their electrochemical gradient)
reversible, can hydrolyze ATP to pump protons in reverse if needed
contains multi-subunit rotary motor: F0 domain (proton channel embedded in membrane), F1 domain (catalytic ATP synthesis)
similar structure to V-type ATPase
F-type ATPase in Mitochondria
proton enters half channel 1, binds Asp-61 on C subunit (Asp becomes neutral and can enter bilayer)
neutral Asp allows c-ring rotation, moving the next Asp61 into position
Arg-210 interacts to guide proton down half channel 2 → proton exits
C-ring rotates, repeating the cycle
rotating C subunit is driven by proton gradient
F-type ATPase in Mitochondria FIGURE