H2Bio: DNA & Genomics 1+2
1/56
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
57 Terms
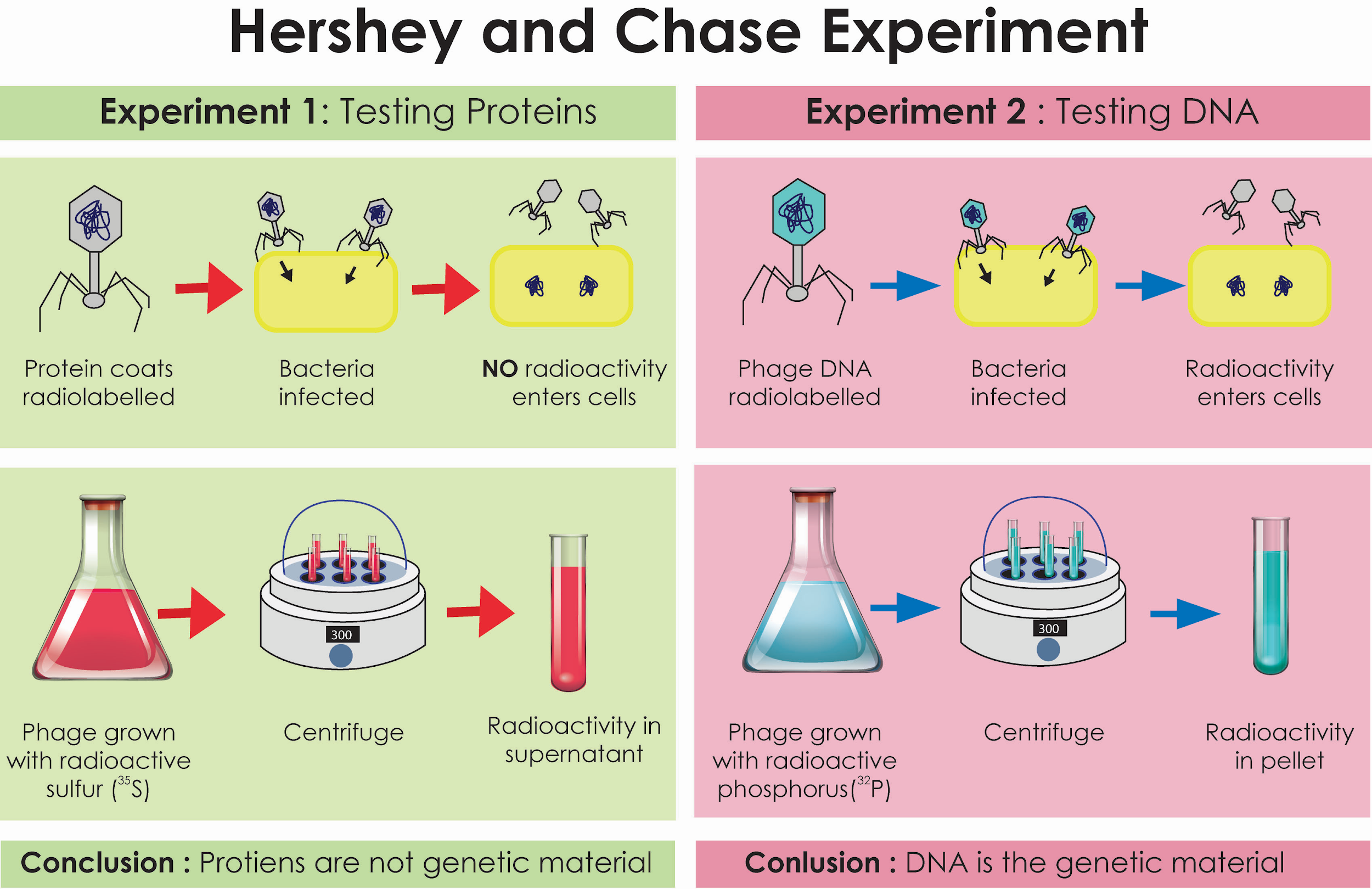
Experiment name to prove DNA is the genetic material
Hershey Chase Experiment: use of radioactive bacteriophages (expt 1 with radiolabelled viral protein coats, expt 2 with radiolabelled viral dna) to infect bacterial cells to observe if radioactivity was transferred to bacterial cells.
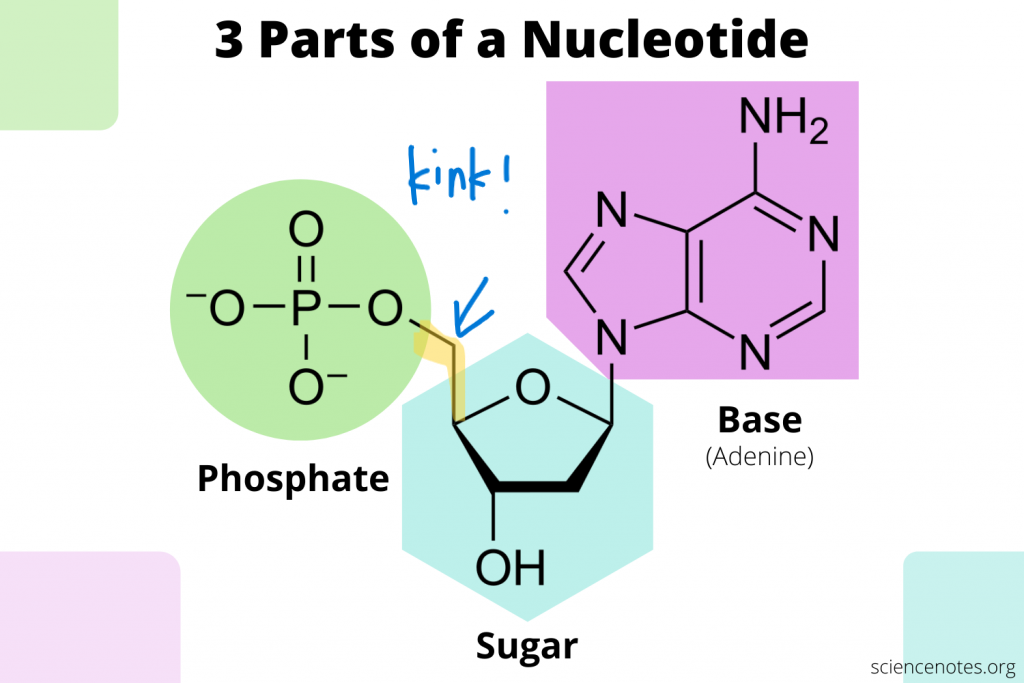
Components of nucleotides (3 components)
pentose/5-carbon sugar (DNA: deoxyribose sugar, RNA: ribose sugar)
nitrogenous base (DNA: A=T, C- - -G) (RNA: A=U, C- - -G)
phosphate group
nucleoside =
nitrogenous base attached to a pentose sugar (without phosphate group)
N base is attached to which carbon by what bond
C1, covalent bond
phosphate grp is attached to which carbon by what bond
C5, phosphodiester bond
where does the next nucleotide attach in forming a long DNA/RNA or to form the phosphodiester backbone?
3’OH group
How do ribose and deoxyribose sugars defer and where?
Ribose has an -OH group at the 2' carbon, while deoxyribose has an -H at the same position.
Purines: definition and 2 examples (babies)
Bases with the 2 ring structures. (babies got 2 eyes) Guanine and Adenine. (babies go GAGA)
Pyrimidines: definition and 3 examples
Bases with the single ring structure. Cytosine, Uracil, Thymine (pyramids CUT)
Chargaff’s rule (in DNA)
no. of purines = no. of pyrmidines
A:T = 1:1, C:G = 1:1 by complementary base pairing
What is complementary base pairing
the formation of weak hydrogen bonds between purines (G or A) and their corresponding pyrimidines ()
RNA is typically single-stranded, how do double-stranded regions of RNA occur?
RNA may sometimes fold back onto itself.
DNA is antiparallel: meaning…
one DNA strand runs in the 5’ to 3’ direction while the other strand runs in the 3’ to 5’ direction. —> DNA has directionality.
Dimensions of DNA: width of sugar phosphate backbone
DNA has a constant width of 2.0nm = width of one purine + one pyrimidine
Dimensions of DNA: distance of each turn of double helix and how many base pairs it contains?
3.4nm = 10 base pairs
DNA vs RNA characteristics (3)

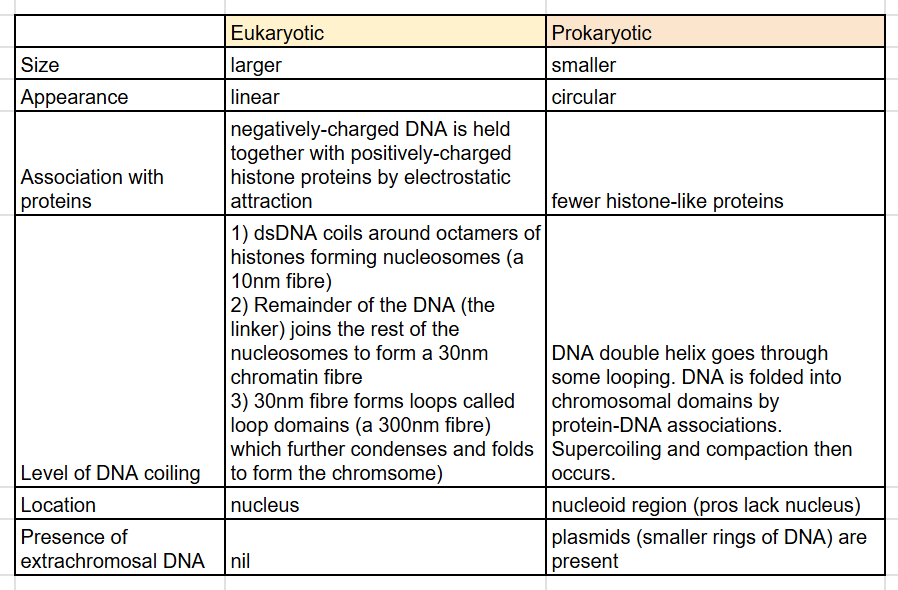
structure of prokaryotic vs eukaryotic genomes (6)
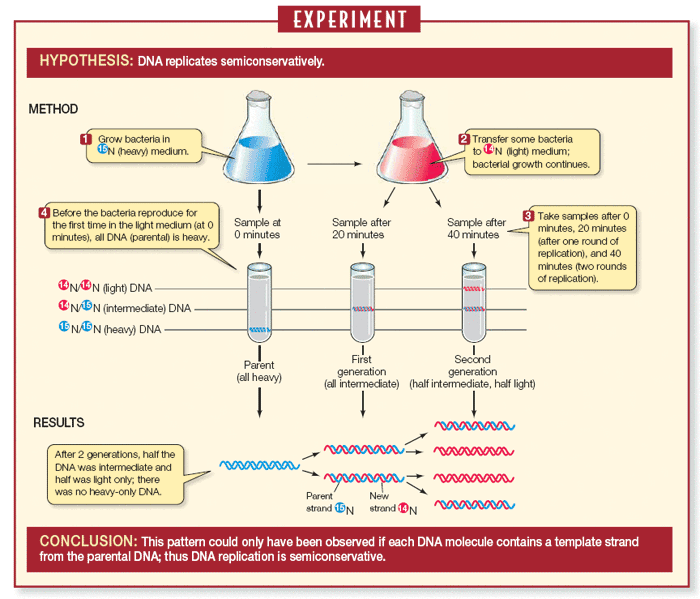
Experiment name for proof of semi-conservative DNA replication
Meselson-Stahl experiment.
1) a stock of parental E. coli was grown for many generations in a 15N medium as the only source of nitrogen until 15N was incorporated into the nitrogenous bases of all the parental E.coli DNA.
2) The E. coli containing 15N-15N was transferred to a medium containing only 14N. transferred E. coli was then allowed to divide once and were collected. The DNA was centrifuged and extracted in CsCl was all hybrid (14N-15N)
3) Some of the hybrid cells were allowed to divide in the 14N-14N medium once again, resulting in half 14N-14N DNA and half 14N-15N DNA.
Hypothesis for conservative DNA replication
2 parental strands reassociate after acting as templates, thus restoring the original double-helix. Newly-synthesised daughter DNA molecules consists only of 2 newly-synthesised strands.
Hypothesis for dispersive DNA replication
Parental DNA are fragmented and dispersed, daughter DNA molecule is made up of a mixture of old and newly synthesised parts.
Hypothesis for semi-conservative DNA replication.
both strands separate by the breaking of hydrogen bonds between the parental strands. both strands are used as template for the synthesis of a new strand through complementary base pairing (a=t, c- - - g). Each newly synthesised strand is a hybrid strand that consists of a parental strand and a newly synthesised strand.
naming of nucleotides as nucleosides
adenine deoxyribonucleotide (that has 3 phosphates) = deoxyirboadenine triphosphate (dATP)
DNA replication occurs in the _____ phase of ______. Where do the building blocks come from?
S phase of interphase. Free deoxyribonucleoside triphosphates are synthesised in the cytoplasm and enter the nucleus vis nuclear pores.
DNA Replication Initiation: Helicase, single-strand binding proteins, topoisomerase
1) DNA replication begins at the origin of replication. Helicase binds to the origin of replication and disrupts the hydrogen bonds between the parental DNA strands, causing them to unzip and separate. (process requires ATP)
2) Single-strand binding proteins bind to each parental strand, keeping them separated such that they can act as templates for DNA replication.
3) Topoimerase relieves ‘overwinding” strain ahead of replication forks by breaking, swivelling and rejoining DNA strands.
DNA replication: Elongation: Primase, RNA Primer, DNA Polymerase iii, DNA polymerase i, DNA Ligase
1) RNA primer is added to each parental strand by primase, providing a free 3’OH end for DNA Polymerase III to attach free deoxyribonucleoside triphosphates to.
2) DNA Polymerase III uses the parental strand as a template to align activated free deoxyribonucleoside triphosphates in a sequence complementary to the template strand.
3) adenine base pairs to thymine with 2 hydrogen bonds and vice versa. cytosine base pairs to guanine with 3 hydrogen bonds and vice versa.
4) DNA polymerase catalyses the formation of phosphodiester bonds between the adjacent daughter DNA nucleotides of the newly synthesised daughter strand.
5) As DNA polymerase moves along the template, it proofreads the previous region for proper base pairing. Any incorrect nucleotide is removed and replaced by the correct one.
6) The leading strand is synthesised continuously in the 5’ to 3’ direction.
7)The lagging strand is synthesised discontinuously, synthesised in fragments called Okazaki fragments initiated by a different RNA primer each.
8) DNA polymerase I removes the RNA primers and replaces them with a deoxyribonucleotide. DNA ligase seals the nicks between the Okazaki fragments in the lagging strand.
DNA replication: Termination
Complementary daughter and parental strands rewind to form a double helix. Each resultant helix consists of one parental strand and one newly-synthesised daughter strand.
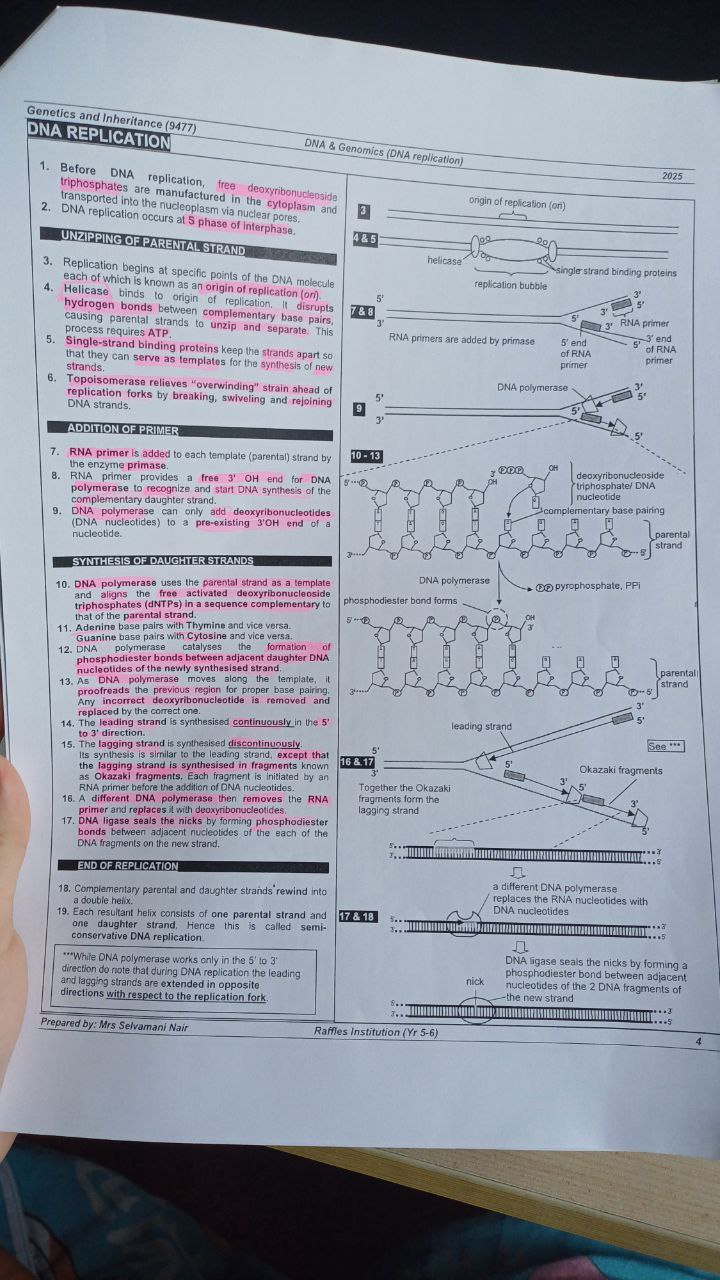
Replication bubble occurs when
there are multiple origins of replication, to speed up the process of DNA replication
main role of DNA
to store genetic information, to pass it from one generation to the next
Why is DNA a suitable store of information: Ru of RuBIcS
Coded information is readily utilised.
weak hydrogen bonds allows template and non-template strand to be separated easily, allowing strands to be used as templates for transcription.
complementary base pairing allows faithful transfer of information during transcription from DNA to RNA, which is then translated to proteins
Why is DNA a suitable store of information: B of RuBIcS
There is a backup of the code.
there are 2 strands in DNA
if one strand is mutated, the other strand acts as the template for the repair of the other
Why is DNA a suitable store of information: Ic of RuBIcS
Daughter cells have identical copies of DNA as the parent cells.
Weak hydrogen bonding allows for parental strands to be separated and used as template strands for new strand synthesis.
Parental strands act as templates of synthesis of new strands by complementary base pairing to produce daughter DNA molecules that are genetically identical (A forms 2 H bonds with T, C forms 3 H bonds with G)
Why is DNA a suitable store of information: S of RuBIcS
DNA is a stable molecule.
numerous H bonds holding the 2 strands together
Adjacent nucleotides in each strand are joined together by strong phosphodiester bonds
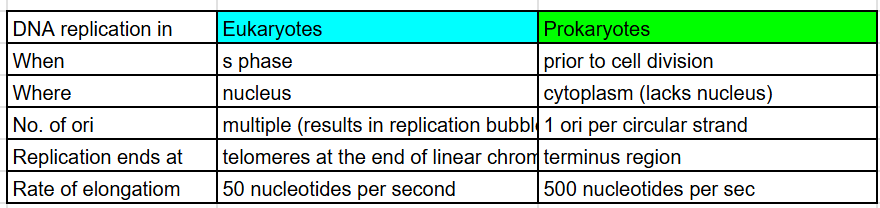
Replication in eukaryotes vs prokaryotes
End Replication problem caused by:
occurs in linear eukaryotic DNA, 3’ Overhang of the parental molecule paired with the lagging strand (as lagging strand lacks a 3’OH end to add a deoxyribonucleotide to replace the initial RNA primer)
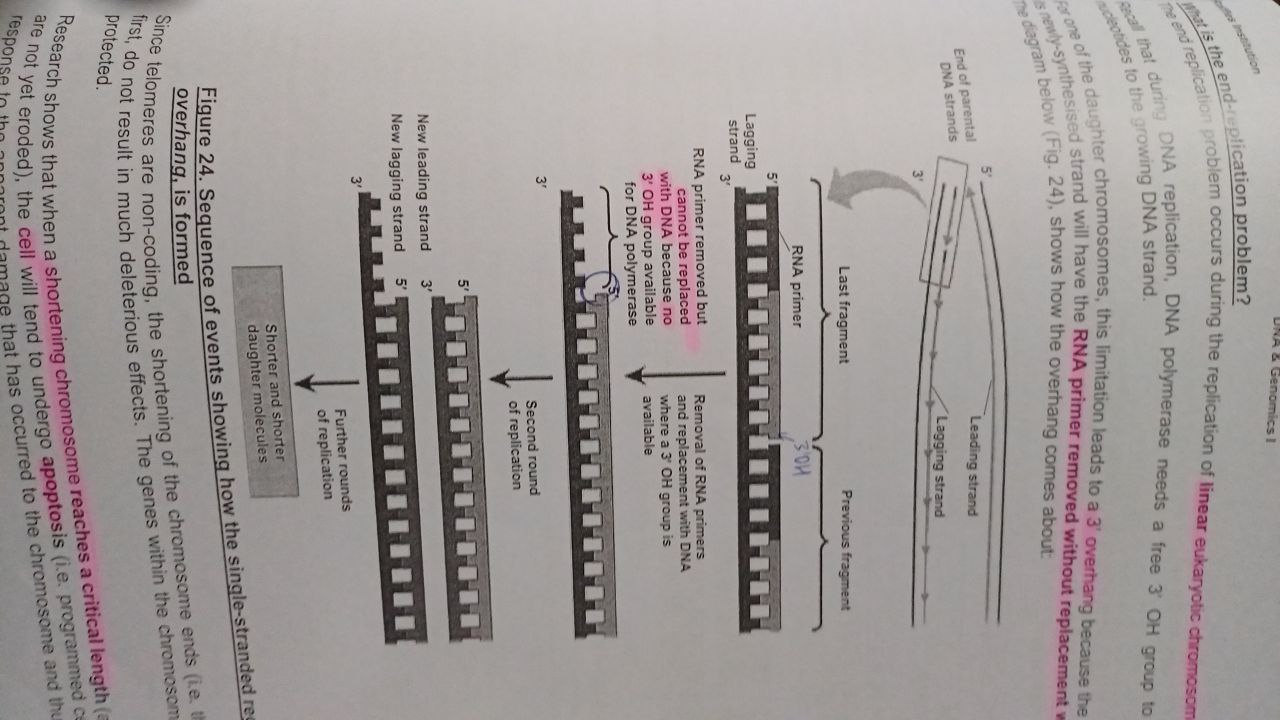
What are telomeres? what is the sequence in humans?
non-coding regions of DNA made up of a series of tandem repeat sequences. Humans: 5’ TTAGGG
Function of telomeres? What happens when telomeres are sufficiently eroded?
Ensures genes are not eroded, prevents loss of genetic material. When telomeres are sufficiently eroded and DNA reaches critical length (but still before any genes can be eroded), cell undergoes apoptosis.
In what cells do telomeres get extended? By which enzyme?
Telomeres are extended by telomerase in stem cells, allowing stem cells to divide infinitely.
Gene definition
specific sequence of nucleotides in a DNA molecule, that code for a specific sequence of amino acids in one polypeptide chain
what is the central dogma? what are the 3 central processes involved?
the theory that genetic information flows from DNA to RNA to proteins (through replication, transcription and translation/protein synthesis)
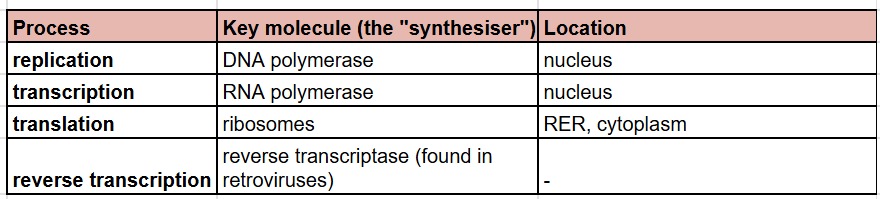
replication VS transcription VS translation VS reverse transcription (key molecule and location)
different permutations of the genetic code lead to how many different kinds of amino acids
20 types
why is the genetic code a triplet code?
If 1 nucleotide codes, there are 4 bases = 4 different codes = 4 amino acids (a.as)
if 2 nucleotides code, there are 4×4 = 16 different codes = 16 a.as
if 3 nucleotides code, there are 4×4×4 = 64 different codes = 64 a.as
Characteristics of the genetic code (6)
triplet code (3 nucleotides form a codon), DNA is read in 3 nucleotides at a time (i.e. the reading frame)
the genetic code is degenerate (different codons code for the same a.a.)
the genetic code is continuous
the genetic code is non-overlapping
the genetic code is universal (codes for the same a.a. in every living thing)
the genetic code has a start (AUG) and stop (UAG, UAA, UGA) codon (Are U Gone) (U Are Annonying, U Are Gone, U Go Away)
what is the start codon? what does it code for? what is its function?
AUG (Are U Gone)
Amino Acid: Met
Codes for ribosomes to begin translating mRNA
what are the stop codons? what do they code for?
UAA, UGA, UAG (U Are Annonying, U Go Away, U Are Gone).
they code for the termination of the polypeptide chain in translation
How does genetic information move from inside the nucleus to the cytoplasm?
Genetic information is carried along the DNA in the nucleus.
mRNA is transcribed in the nucleus —> leaving the nucleus through nuclear pores to cytoplasm
Ribosomes interact with mRNA and tRNA to translate information on the mRNA into a polypeptide.
TRANSCRIPTION definition
process in which the nucleotide of a gene is used as a template for the synthesis of RNA made up of complementary sequences.
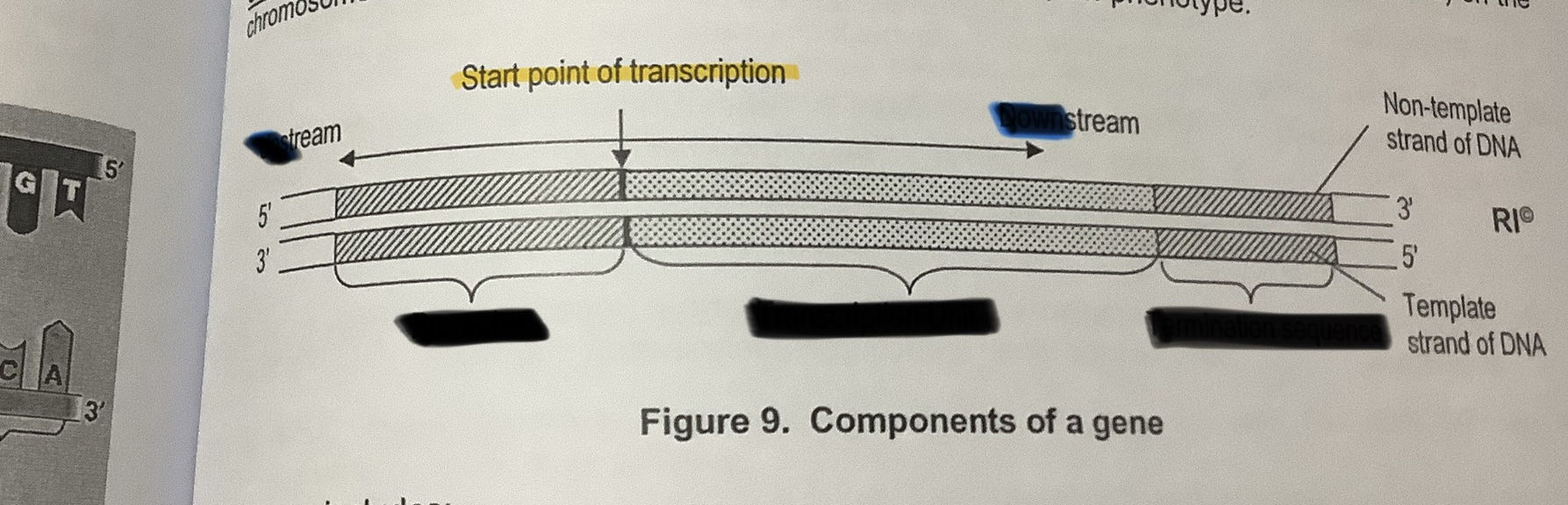
Label the components of a gene
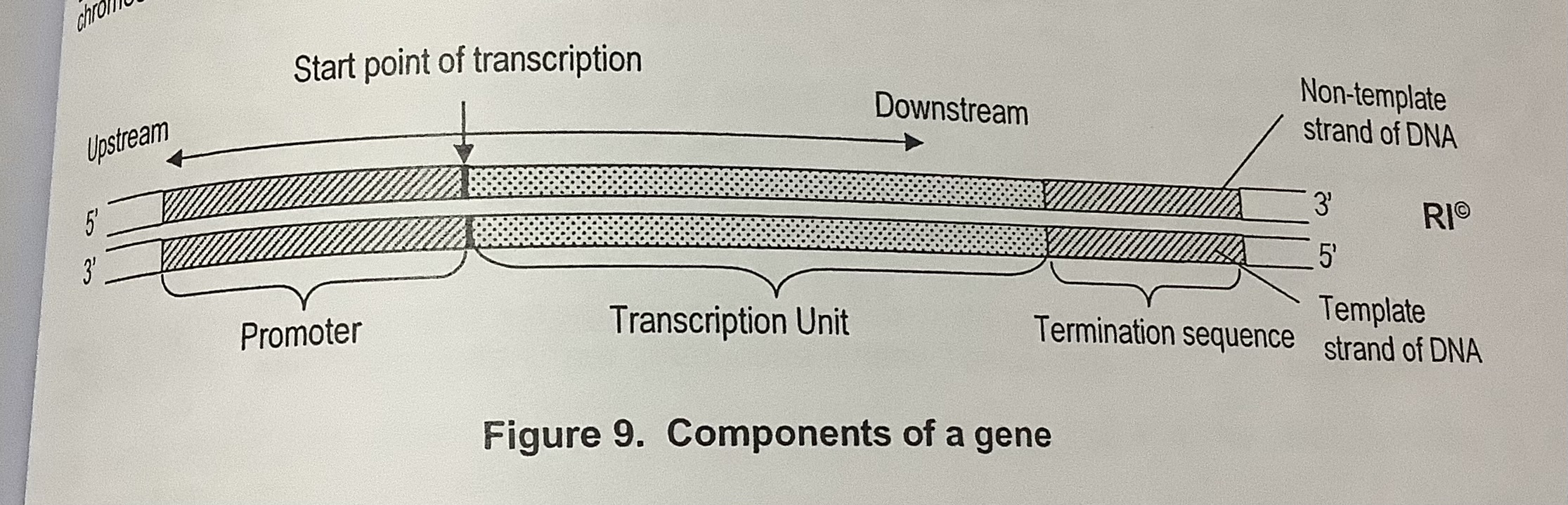
strands are named the ______ strand and _________ strand respectively
Template strand
Non-template strand or Coding strand
TRANSCRIPTION: Initiation
General Transcription Factors (GTF) will recognise and bind to the promoter
GTFs will recruit RNA polymerase and position it at the promoter
RNA polymerase & GTFs form the transcription initiation complex
RNA polymerase will unzip the DNA double helix by disrupting the H bonds between the complementary base pairs
Only one strand is used as the template to synthesise a complementary mRNA strand
TRANSCRIPTION: Elongation
1) Free ribonucleotides undergo complementary base paring by deoxyribonucleotides on the template DNA strand.
2) Adenine, cytosine, guanine and uracil- containing ribonucleotides complementary base pair with thymine, guanine, cytosine and adenine-containing deoxyribonucleotides on the template DNA strand.
3) Cytosine forms 2 hydrogen bonds with guanine; adenine forms 3 hydrogen bonds with thymine and uracil.
4) RNA polymerase catalyses the formation of phosphodiester bonds between adjacent ribonucleotides, forming the sugar-phosphate backbone.
5) mRNA is synthesised in the 5’ to 3’ direction by adding free ribonucleotides to the free 3’OH end
6) While the transcription complex moves down the template DNA double helix, the regions that have just been transcribed are reannealed.
TRANSCRIPTION: Termination
After RNA polymerase transcribes through the termination sequence, pre-mRNA is released and the RNA polymerase will dissociate, terminating transcription. A sequence known as a polyadenylation signal (AAUAA) will be present in the pre-mRNA.
Post-transcriptional modification occurs in which karyotes? why?
Occurs in eukaryotes.
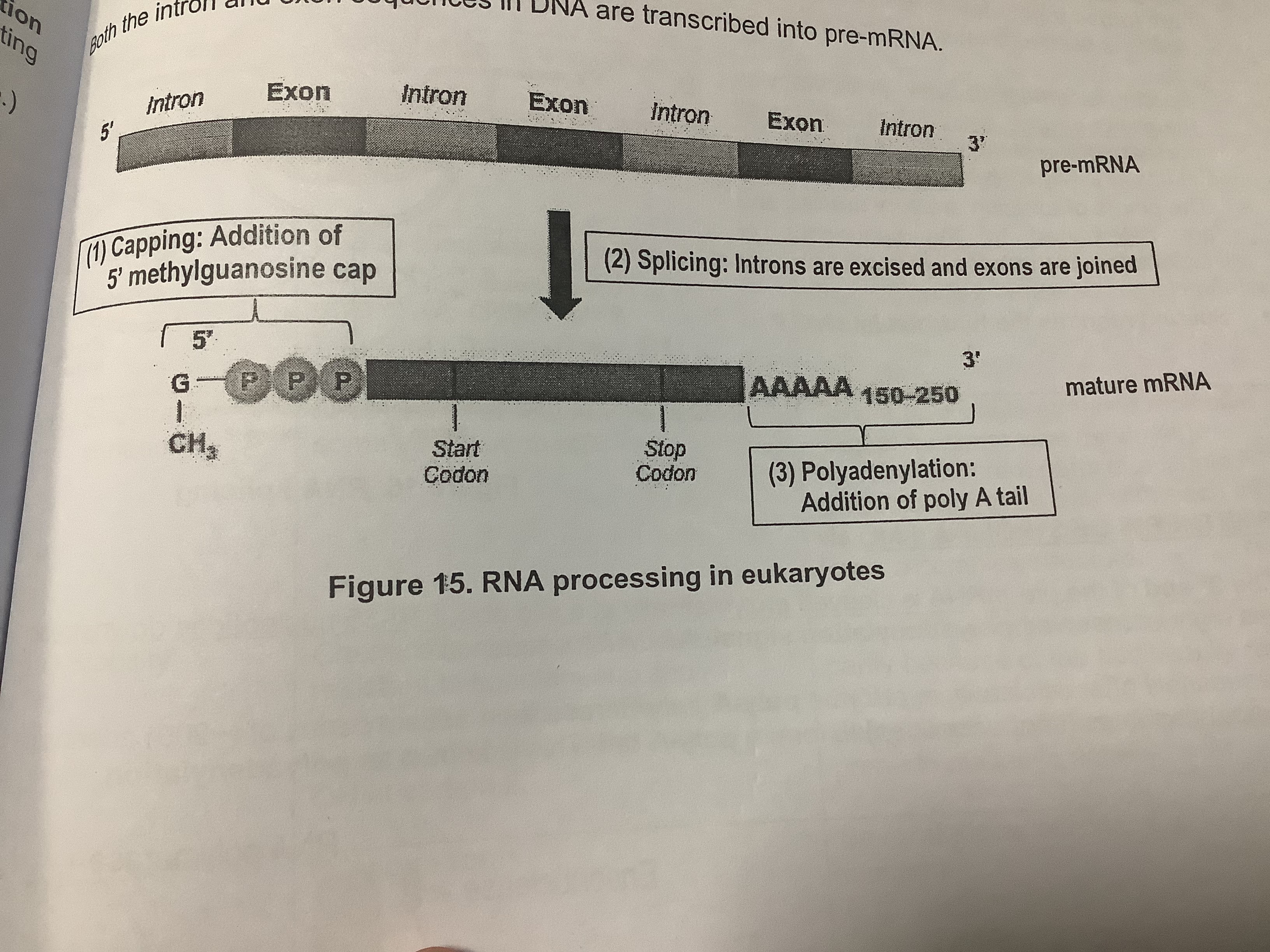
3 post-transcriptional modifications:
1) Addition of a 7-methylgunanosine cap to the 5’ end of pre-mRNA: protects mRNA from degradation by ribonucleases, facilitates the export of mature mRNA from the nucleus to the cytoplasm as it helps recognise mRNA so that it can undergo subsequent steps such as splicing
2) RNA splicing: spliceosomes excise introns (non-coding regions) and join exons (coding regions)
3) Synthesis of a poly A tail (polyadenylation): protects mRNA from degradation by ribonucleases, facilitates the export of mature mRNA from the nucleus to the cytoplasm
Function of ribonucleases
To degrade mRNA that is faulty or damaged to regulate gene expression in the cell.
role of mRNA
1) act as the template of translation
each codon on the mRNA will determine the polypeptide sequence
2) takes information out of the nucleus through the nuclear pores to the cytoplasm where translation occurs
role of tRNA