bio 305 exam 4
1/241
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
242 Terms
pre-discovery of mRNA
pulse-chase method was used to track newly synthesized RNA, using radioactive nucleotides
Volkin and Astrachan studied transcription in bacteria after infection by a bacteriophage. . .they saw that radioactivity from uracil broke down quickly, so translation involves a type of RNA with short lifespan
subsequent experiments also showed that radioactivity was concentrated in the nucleus and lingered for a short time in the cytoplasm, indicating that the RNA was likely an intermediary
discovery of mRNA
Brenner, Francois, Jacob, and Meselson did an experiment to determine whether new phage protein synthesis in bacteria needed newly constructed ribosomes or existing bacterial ribosomes
experiment found that existing ribosomes are used to produce phage proteins, and RNA that directed this protein synthesis formed and degraded quickly
this allowed them to conclude that phage “messenger” RNA with a short half-life is responsible for protein synthesis during infection
snRNA
small nuclear RNA, found in nucleus of eukaryotic cells, participate in mRNA processing and intron removal
miRNA and siRNA
recently recognized types of regulatory RNA that are active in animal and plant cells, important for controlling stability or translatability of certain mRNAs. . .implicated in gene regulation
4 stages of transcription
promoter recognition and identification
initiation of transcript synthesis
transcript elongation
transcript termination
upstream
near 5’ start of transcript
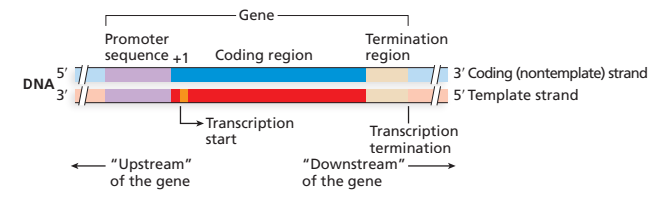
downstream
near 3’ end of transcript
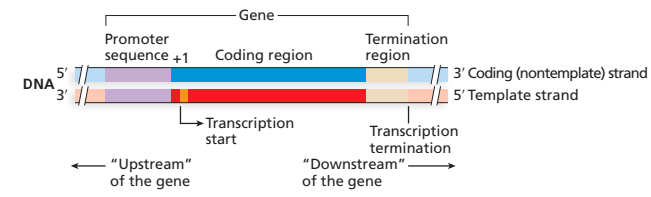
promoter
nucleotide sequence that’s not transcribed, a transcription-regulating DNA sequence that controls access of RNA polymerase to the gene
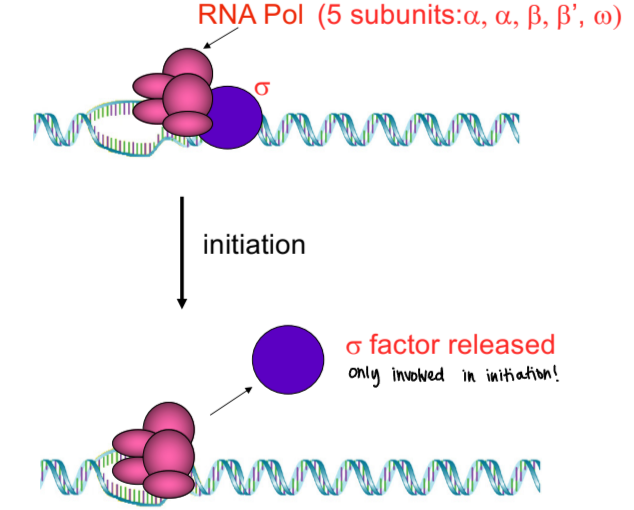
bacterial RNA polymerase
only one bacterial RNA polymerase (E. coli), came from experiment with antibiotic rifampicin
five-polypeptide RNA polymerase core that binds to 6th polypeptide (sigma subunit) that has a conformational change in core enzyme into active form (holoenzyme)
sigma subunit allows core enzyme to bind specifically to promoter sequences
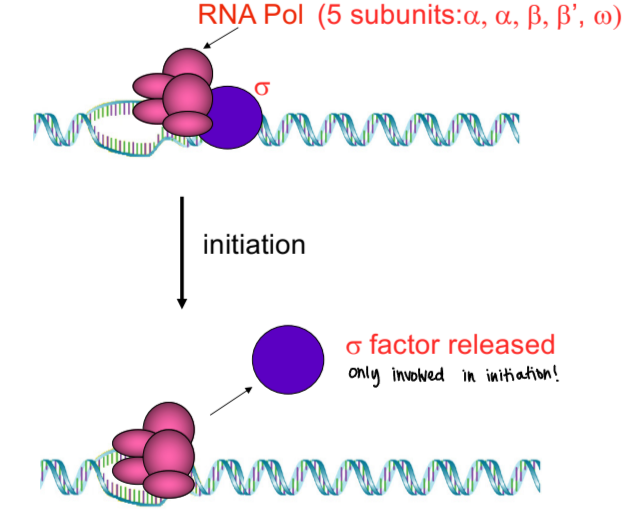
sigma factor
RNA pol subunit in prokaryotes that is only involved in initiation
alternative sigma subunits
alter specificity of holoenzymes for promoter regions by imparting distinct conformational changes to the core
consensus sequences
short regions of DNA sequences that are highly similar, though not necessarily identical
structure of prokaryotic protein coding genes and mRNAs
DNA
—(-35)———(-10)—(+1, transcription start)———————————————(terminator sequence)
mRNA
(5’ UTR)[start codon]———coding sequence———[stop codon](3’ UTR)
![<p>DNA</p><p>—(-35)———(-10)—(+1, transcription start)———————————————(terminator sequence)</p><p></p><p>mRNA</p><p>(5’ UTR)[start codon]———coding sequence———[stop codon](3’ UTR)</p>](https://knowt-user-attachments.s3.amazonaws.com/f58d95e8-eb73-45c1-a136-2f604268e772.png)
pribnow box sequence
-10 consensus sequence consisting of 6 bp 5’ TATAAT 3’ and separated by 25 bp from another 6 bp region (-35) 5’ TTGACA 3’
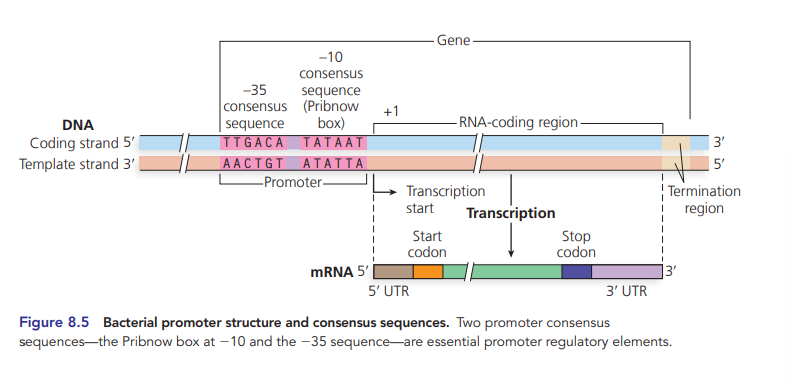
UTR
untranslated mRNA
how does RNA polymerase holoenzyme initiate transcription? (prokaryotes)
initial loose attachment to double-stranded promoter sequence and then binds tightly to it to form closed promoter complex
bound holoenzyme unwinds about 18 bp of DNA around -10 consensus sequence to form open promoter complex
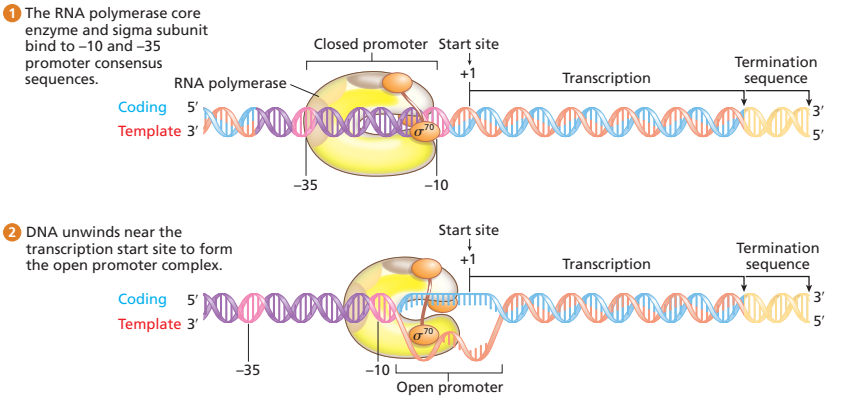
transcription elongation
when the holoenzyme reaches 1+ nucleotide, it beings RNA synthesis using the template strand
holoenzyme remains intact until first 8-10 nucleotides have joined, then sigma subunit dissociates from core enzyme, which keeps on going. . . sigma subunit can go to another core enzyme to transcribe another gene
DNA is unwound and then rewound after the enzyme passes
end product of transcription
ssRNA that’s complementary and antiparallel to template DNA strand
transcription termination mechanisms
usually signaled by DNA termination sequence containing repeating sequence producing distinctive 3’ RNA sequences
intrinsic and rho-dependent termination (less frequent)
intrinsic termination
two features: inverted repeat and string of adenines in template DNA beginning at 5’ end of inverted repeat 2 region (3’ end of mRNA)
transcription of inverted repeats makes mRNA with complementary segments that fold into short ds stem ending (hairpin)
string of uracils complementary to adenines follow haripin structure on 3’ end of mRNA
those things cause RNA polymerase to backtrack to hairloop and destabilize, until it falls off DNA and transcript is released
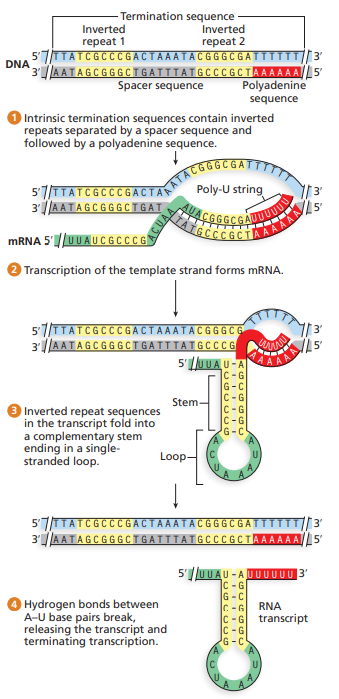
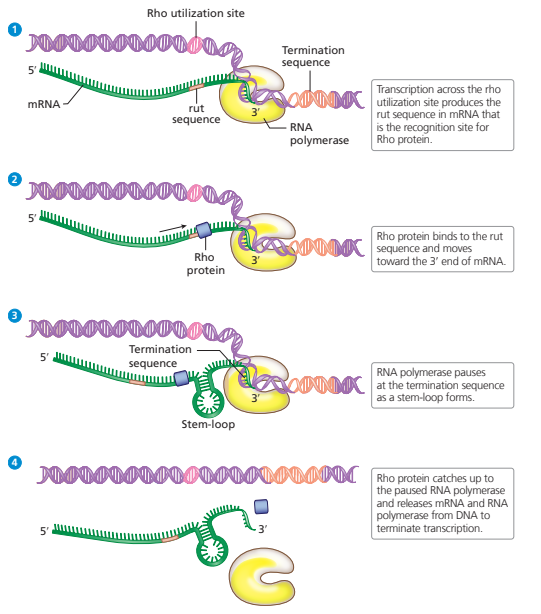
rho-dependent termination
less common, requires activation of rho protein to bind to new mRNA and catalyze separation of mRNA from RNA polymerase to terminate transcription
rho utilization site transcription produces rut site on mRNA, where rho protein attaches and moves towards RNA polymerase
when RNA polymerase reaches terminator sequence hairpin, rho protein can catch up and catalyze release
order of information flow, aka the “basic assumption”
DNA ↔ RNA —> protein
protein is last stage of flow, can’t go back from that
DNA can recreate itself (replication) and so can RNA, protein can recreate itself in some cases (like prions)
dogma
a principle or set of principles laid down by an authority as incontrovertibly true. . .may not always be the case, and could be detrimental in a science. . . Dr. Wierzbicki calls them the enemy of knowledge
unique quality of RNA
its function and structure is in between DNA and proteins. . .
it can carry genetic info in nucleotide sequence like DNA
can form complex structures and carry out biochemical functions, like proteins (can act as enzymes)
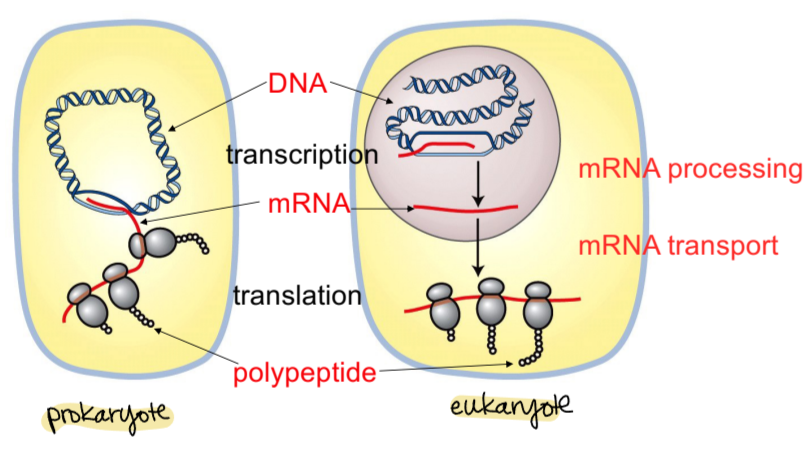
compare and contrast prokaryote and eukaryote transcription/translation
prokaryotes:
circular genome
DNA transcribed to mRNA and mRNA translated to proteins at same time
eukaryotes:
linear genome
DNA transcribed into mRNA in nucleus
mRNA processed and transported
mature mRNA translated to proteins in cytoplasm
three RNA polymerases in eukaryotes
RNA pol I, RNA pol II, RNA pol III
RNA polymerase I
transcribes several ribosomal RNA genes
RNA polymerase II
transcribes mRNA that encodes polypeptides, also snRNA
RNA polymerase III
transcribes all tRNA genes and one snRNA and rRNA gene
memorization hack for eukaryotic RNA polymerases: RooMaTe
RooMaTe
R is first in mnemonic, RNA polymerase I is for rRNA
M is second in mnemonic, RNA polymerase II is for mRNA
T is third in mnemonic, RNA polymerase III is for tRNA
some exceptions. . . RNA pol II and III both make snRNA also, and RNA pol III also makes one rRNA gene
structure of eukaryotic protein coding genes and mRNAs
DNA
—(TATA-box)—(+1, transcription start)————————————————(polyA signal)
mRNA
cap(5’ UTR)[start codon]———coding region———[stop codon](3’ UTR)AAAAA
![<p>DNA</p><p>—(TATA-box)—(+1, transcription start)<mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark><mark data-color="yellow" style="background-color: yellow; color: inherit">—</mark><mark data-color="blue" style="background-color: blue; color: inherit">—</mark>(polyA signal)</p><p></p><p>mRNA</p><p>cap(5’ UTR)[start codon]———coding region———[stop codon](3’ UTR)AAAAA</p>](https://knowt-user-attachments.s3.amazonaws.com/82d3b3a3-835f-48fe-99d9-5ebe1bd268df.png)
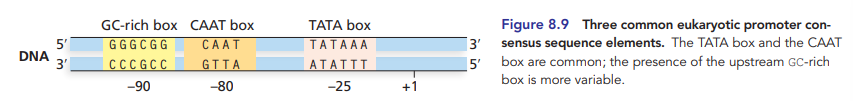
TATA box, aka goldberg-hogness box
most common eukaryotic promoter consensus sequence
approx position -25 to +1 start fo transcription
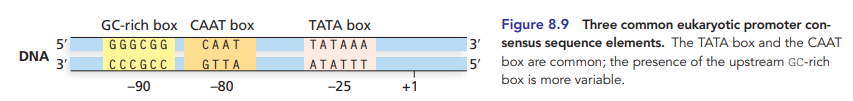
other consensus eukaryotic promoters
CAAT box, GC-rich box, OCT box, etc.
eukaryotic RNA polymerase II holoenzyme
14 subunits, two largest subunits form catalytic site
preinitiation complex for transcription - eukaryotes
composed of RNA pol II, TFIID, general transcription factors (GFTs), and template DNA
general transcription factors (GFTs)
TFIIA, TFIIB, F, E, H, J
very important in telling RNA polymerase where to start transcribing
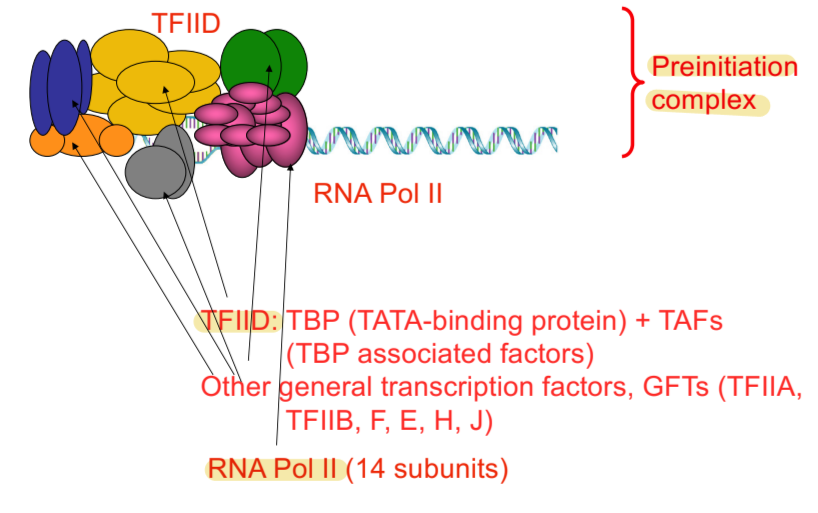
transcription factors
proteins that bind to promoter regulatory sequences and influence transcription initiation by interacting (directly or indirectly) with RNA polymerase II
TFIID
multisubunit protein containing TATA-binding protein (TBP) and subunits of TBP-associated factor (TAF) that binds to TATA box, separates strands of DNA to start transcription
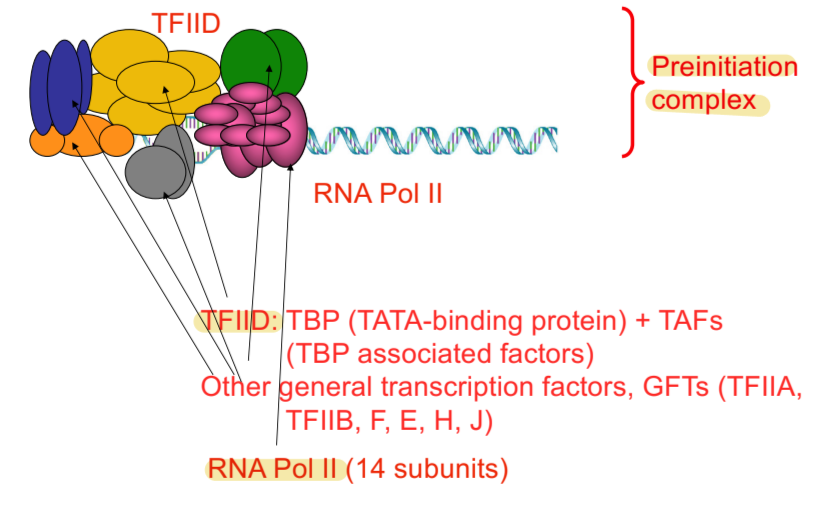
fate of preinitation complex after initiation - eukaryotic transcription
most factors unbind, TFIID remains at promoter to help another RNA pol II molecule, while RNA pol II continues after initiation
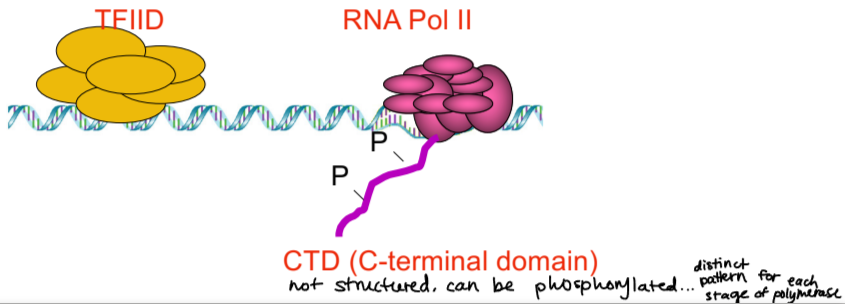
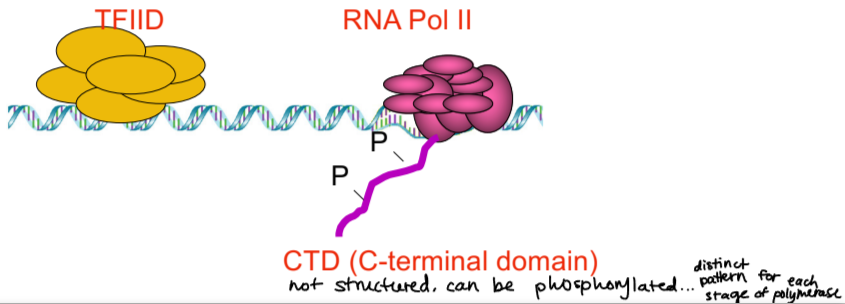
CTD
C-terminal domain
not structured domain
can be phosphorylated, there is a distinct pattern of phosphorylation for each stage of polymerase
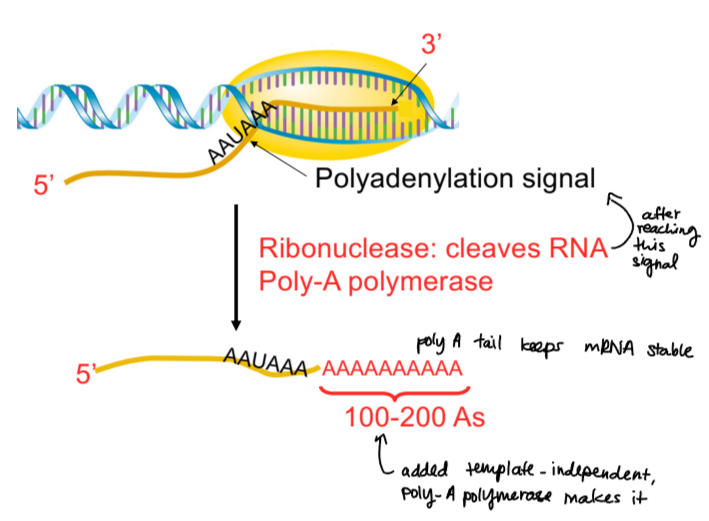
transcription termination in eukaryotes
once RNA pol II reaches polyadenylation signal, a ribonuclease cleaves RNA and poly-A polymerase makes a polyA tail to keep mRNA stable
polyA is template-independent, the polyA tail is not coded for by DNA
RNA processing in eukaryotes
5’ capping, splicing, and poly-adenylation
what coordinates RNA processing?
CTD, through its phosphorylation pattern
cotranscriptional process, no unprocessed RNA floating around
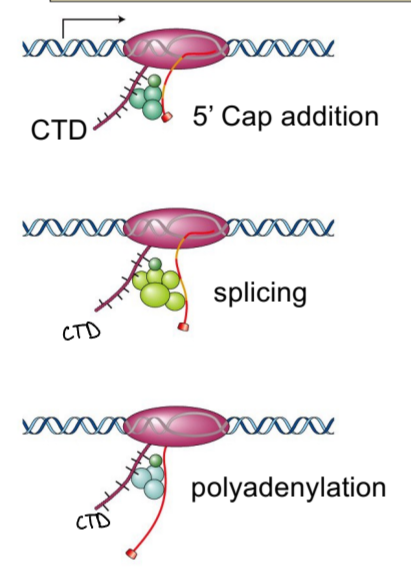
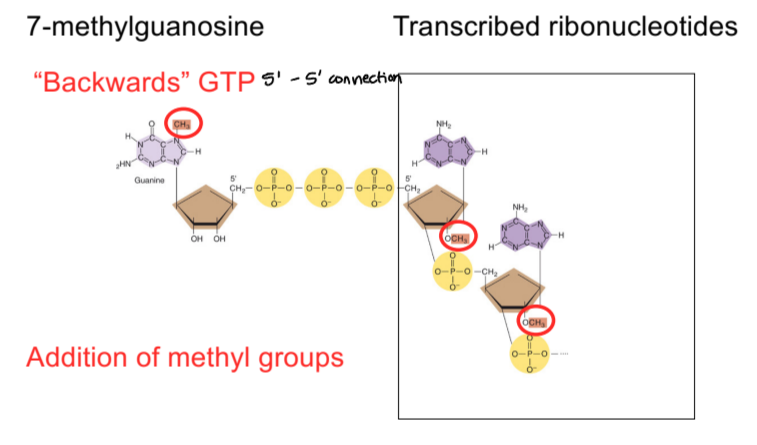
5’ cap
7-methylguanosine, backwards GTP in 5’ to 5’ connection
hnRNA
heterogenous nuclear RNA, immature and unprocessed RNA with exons and introns
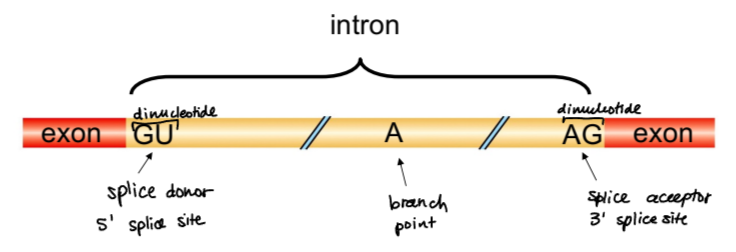
what is RNA splicing?
removal of introns
what is responsible for splicing?
spliceosome, a ribonucleoprotein complex with snRNPs (snRNA and protein) and additional proteins
it can determine splice donor and acceptor sites, we can’t really do that yet
splice donor site
dinucleotide GU, 5’ splice site
slice acceptor site
dinucleotide AG, 3’ splice site
describe the splicing process
spliceosome is assembled
first splicing reaction: cleaving at splice donor site, spliceosome attaches to branch site
second splicing reaction: cleaving at splice acceptor site, exons join and intron removed as lariat, which will eventually degrade
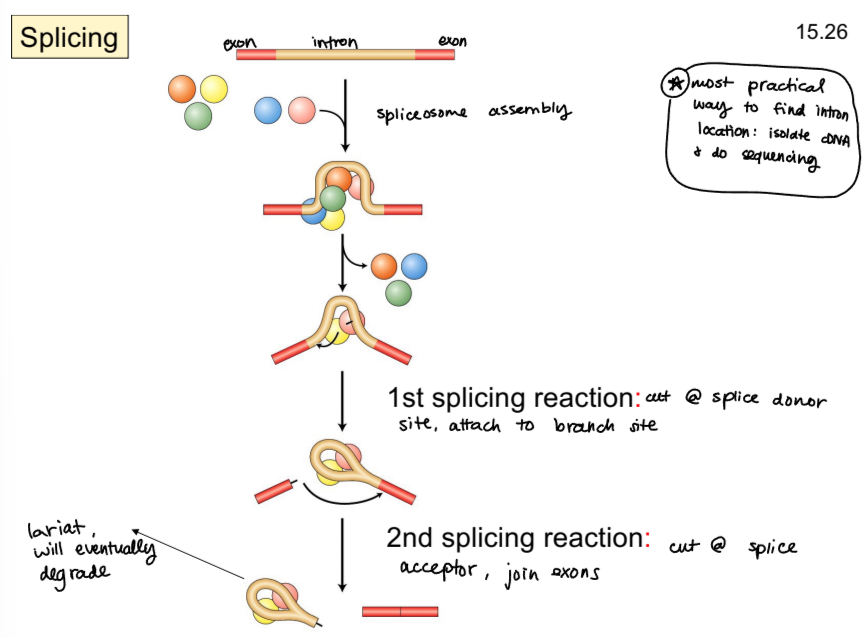
what is the most practical way to find intron location?
isolate cDNA and do sanger sequencing
alternative splicing
one gene can give rise to more than one polypeptide
in human genome, about 25000 genes give rise to about 100000 proteins
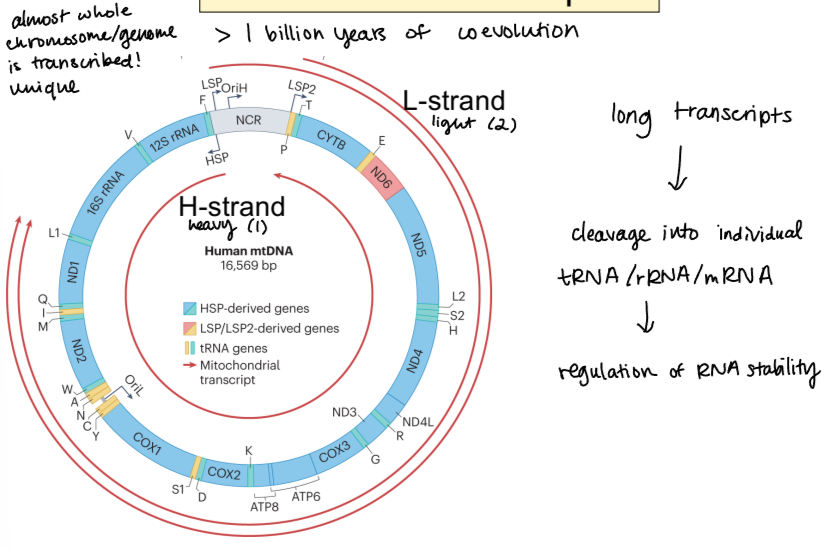
process of mitochondrial transcription
long transcripts that are almost whole genome —> cleavage into individual tRNA, rRNA, and mRNA —> regulation of RNA stability occurs for each fragment individually
what is used for mitochondrial transcription?
phage-type single subunit polymerase (unusual)
3 transcripts produced from mtDNA
H-strand (heavy) and 2 L-strands (light)
snoRNA
small nucleolar RNA, does rRNA processing
what is the function of transcription initiation?
defining the 5’ end of mRNA
enhancer sequences
DNA regulatory sequences that increase level of transcription of specific genes. . .usually located upstream of genes they regulate but can be downstream as well
silencer sequences
DNA sequences/elements that repress transcription of genes, binding proteins that bend DNA so that genes are hidden from transcription activation by RNA pol II
heterochromatin
densely packed chromosomes
euchromatin
loosely packed chromosomes. . . “u” for uncoiled, or “e” for expressed, meaning transcribable!
features of promoters recognized by RNA pol I
core element, -45 to +20
upstream control element, -100 to -150
both are rich in G and C
function of 5’ cap
protecting mRNA from rapid degradation
facilitating mRNA transport across nuclear membrane
facilitating subsequent intron splicing
enhancing translation efficiency by orientating ribosome on mRNA
polyadenylation signal sequence
AAUAAA
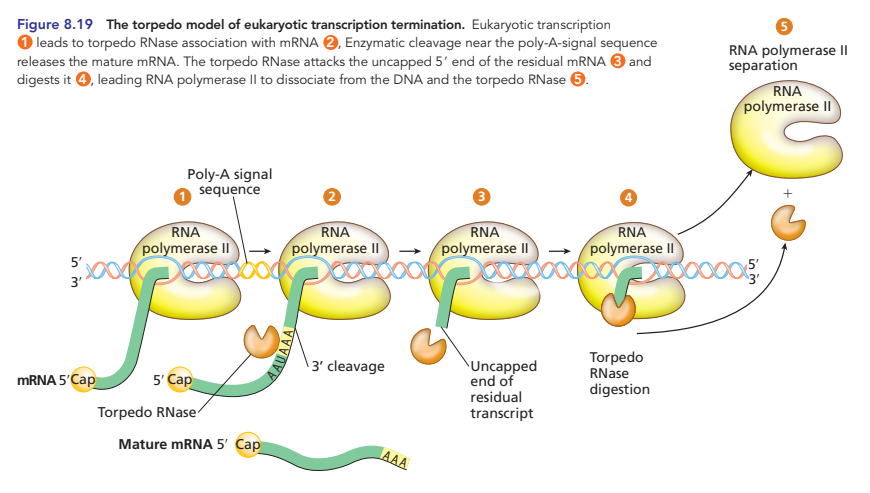
torpedo model of transcription termination
specialized RNase acts as a torpedo aimed at residual mRNA attached to RNA pol II, because it has no cap protecting the 5’ end
it quickly destroys the residual mRNA left after cleaving the mature mRNA from RNA pol II and catches up to RNA pol II, causing it to dissociate from the template DNA and end transcription
branch point
20-40 nucleotides upstream of 3’ splice site, containing a branch point adenine. . .critical for accurate intron removal
self-splicing introns
introns that can self-catalyze their own removal, ribozymes that are classified into group I or group II
rRNA processing
rRNAs are transcribed as large precursor molecules that are cleaved into smaller RNA molecules, denoted in S units (svedberg)
tRNA processing
some produced simultaneously with rRNAs, others transcribed as part of large pre-tRNA transcript that’s cleaved to make multiple tRNAs
nucleotides are trimmed off at 5’ and 3’ ends to prepare
tRNAs fold into 3D structure looking like a clover, with three hairpins and one stem
AA binding site is added to 3’ end (CCA terminus)
what is the direction of translation?
5’ to 3’
corresponding mRNA and protein structural landmarks
N-terminal corresponds to 5’ end, C-terminal corresponds to 3’ end
ribosome tRNA sites
A - accepts new AAs
P - protein polymerizes
E - polypeptide exits ribosome
3 essential tasks of ribosomes
bind mRNA and identify start codon where translation begins
facilitate the complementary base pairing of mRNA codons and tRNA anticodons that determines amino acid order in the polypeptide
catalyze peptide bond formation between AAs during polypeptide formation
ribosome structure
consists of 2 main subunits, large and small, measured in svedberg units (S)
large subunit has enzymatic activity (ribozyme) and has 3 catalytic sites. . . E, P, and A
E. coli ribosome
small subunit is 30S, large subunit is 50S, fully assembled is 70S
mammalian eukaryotic ribosome
small subunit is 40S, large subunit is 60S, fully assembled is 80S
cryo-EM
cryo-electron microscopy, pioneered by robert glaeser in 70s and perfected jaques dubochet in 80s
uses liquid nitrogen or ethane to instantaneously freeze macromolecules and preserve them in their native state
where is the anticodon on tRNA, location where codons bind to?
‘clover’ end directly opposite 3’ end with AA attachment site
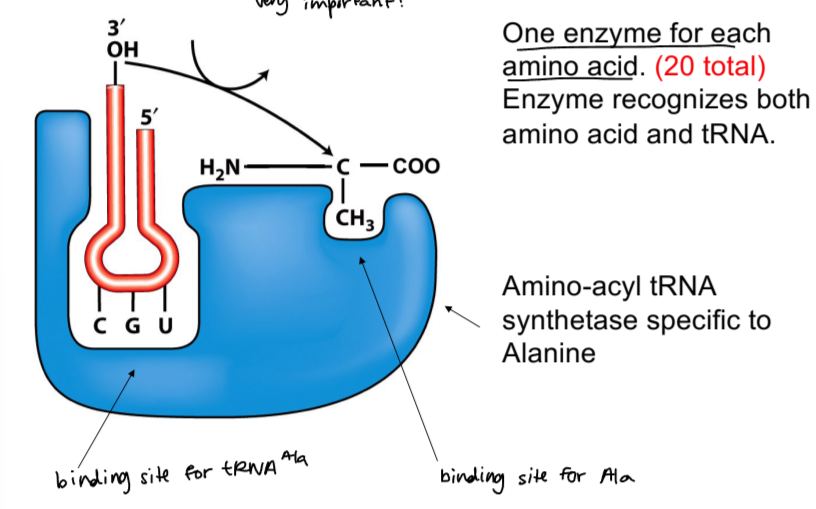
aminoacyl-tRNA synthetase
enzyme specific to each AA (20 total) that recognizes both AA and tRNA
matches correct AA to mRNA codon
translation initiation in prokaryotes
the small subunit, together with initiation factors (IFs) bind the Shine-Dalgarno sequence, near AUG start codon
a charged (AA-bound) tRNA binds
large ribosomal subunit binds
IFs release
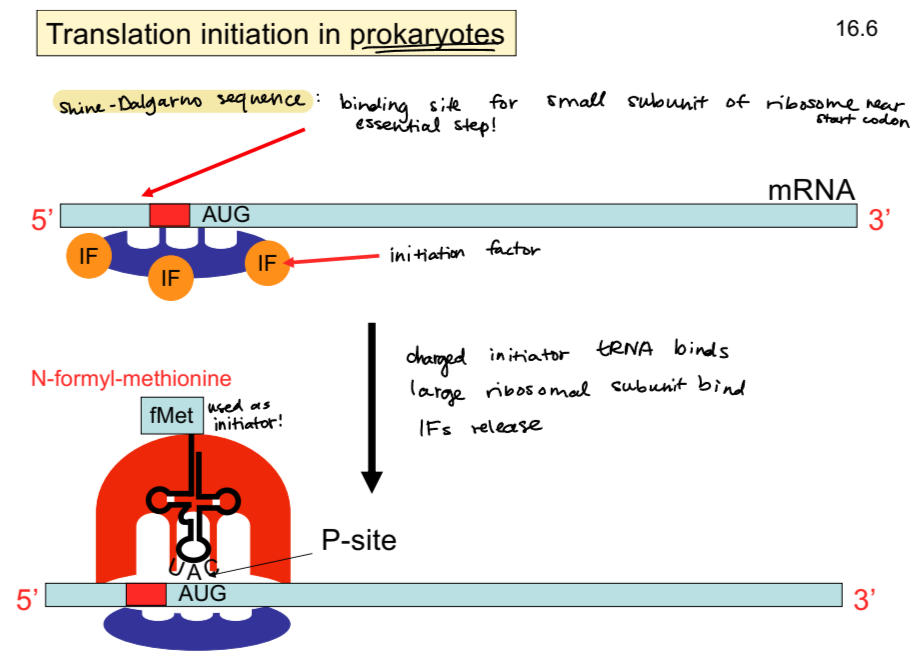
what is the initiator tRNA charged with?
fMet for prokaryotes, Met for eukaryotes
Shine-Dalgarno actual sequence
typically AGGAGGU
translation initiation in eukaryotes
small subunit with eukaryotic initiation factors (eIFs) binds 5’ cap, scans mRNA for AUG start codon
charged initiator tRNA binds
large ribosomal subunit binds
eIFs released
kozak sequence
consensus translation initiation sequence in eukaryotes
5’ ACCAUGG 3’
what provides energy for assembly of ribosome and translation machinery, as well as steps of elongation?
GTP
polycistronic
mRNA in prokaryotes
multiple polypeptides produced from a single transcript due to multiple Shine-Dalgarno sequences and multiple translation initiation sites
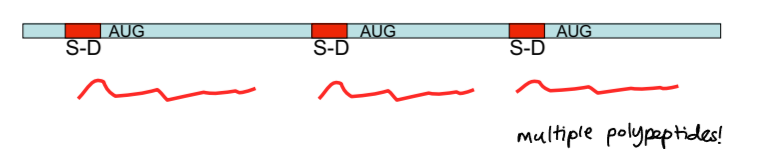
monocistronic
mRNA in eukaryotes
a single polypeptide produced from a single transcript, because the 5’ cap is the only ribosome entry site and there is only one translation initiation site
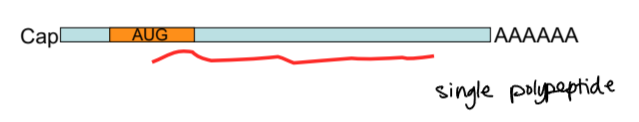
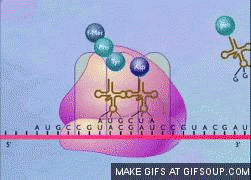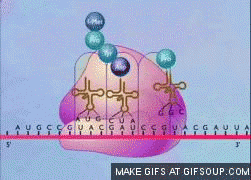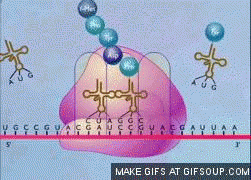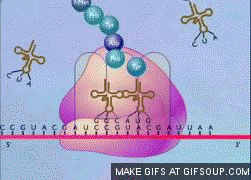
translation elongation steps
similar in prokaryotes and eukaryotes, but eukaryotes have more complex and numerous protein factors
entry of tRNA with bound AA into A site, aided by elongation factor (EF)
ribosome catalyzes peptide bond formation, with newly formed dipeptide in A site
ribosome movement along mRNA causes dipeptide to move to P site and uncharged tRNA from P site moves to E site and is released
repeat!
translation termination steps
release factor (RF) binds stop codon
catalytic activity of RF makes polypeptide dissociate from tRNA
tRNA and mRNA separate from ribosome
ribosome dissociates into large and small subunits
features of genetic code, as demonstrated by experiments
number of codons per AA is variable
three stop codons
universal
no overlaps or gaps between codons
degenerate code, more than one code per amino acid, so the third nucleotide isn’t as important
what can the different genetic codes be understood as?
same information in different languages
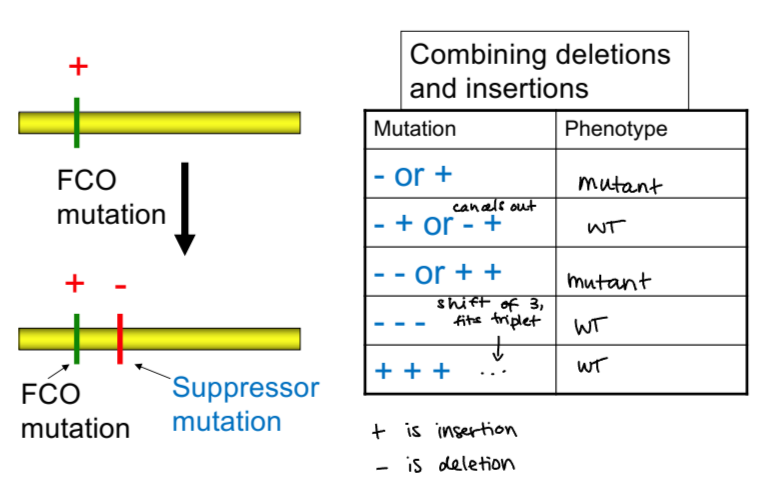
evidence for triplet nature of genetic code - crick and brenner 1955
used proflavin, a molecule that distorts DNA’s double helix and can cause insertions and deletions
created mutations in rII gene
one or two insertions or deletions caused frameshift mutations that change output, but three insertions or 3 deletions (or same number of both deletions and insertions) cause no change
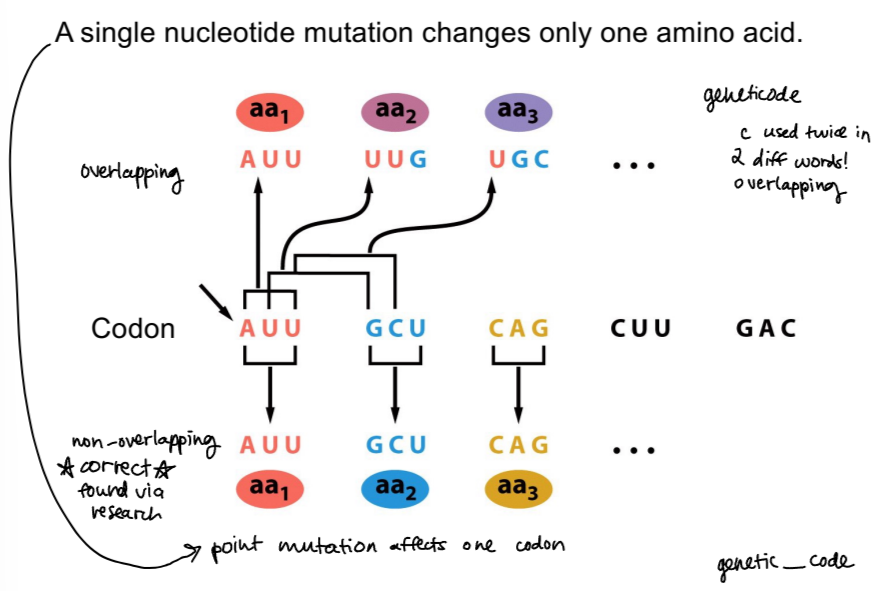
evidence that codons are nonoverlapping - Heinz Fraenkel-Conrat 1960
if overlapping - one alteration can affect 3 consecutive codons. . . if nonoverlapping - one alternation affects one codon
used nitrous oxide, which caused a mutation of a single nucleotide
found that only one codon/amino acid was altered, so codons are nonoverlapping!
how were codons assigned to amino acids? nirenberg, matthaei, khorana
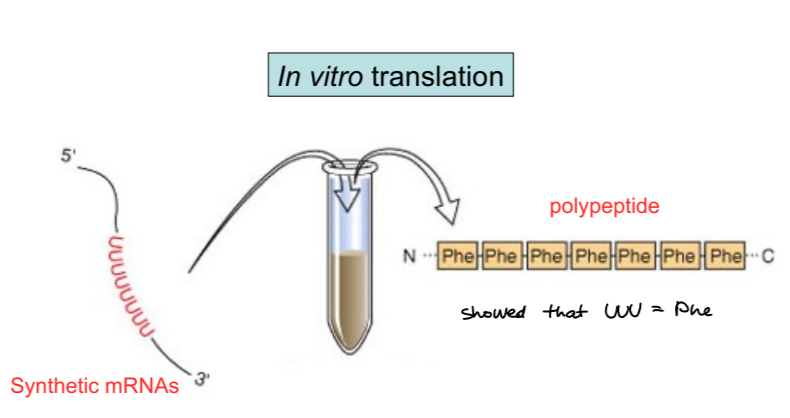
nirenberg and matthaei created synthetic mRNAs with specific sequences (like polyU or polyA) and did in-vitro translation
khorana developed synthetic mRNAs with repeating di-, tri-, and tetranucleotide patterns, which revealed even more
last piece as nirenberg’s experiment with mini mRNAs (one codon) and radioactively labeling one of 20 AAs, and they filtered the mixture to only capture mRNA/tRNA/ribosome complex, and were able to match codon to AA for all combos
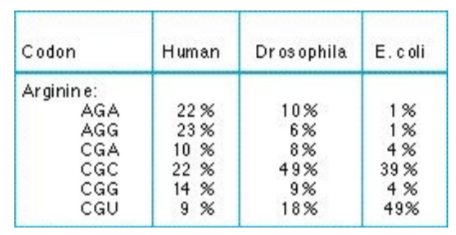
codon bias
frequency of synonymous codons differs in various species
different codons for the same AA have different expression levels across species
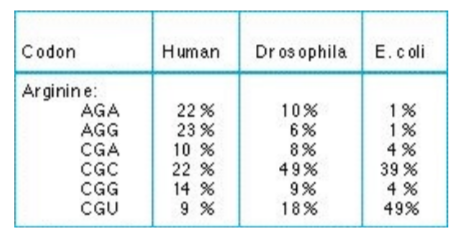
what do you need to do if you want to express a protein from one species in another?
optimize the genetic code to get the highest level of translation! choose synonymous codon with the highest expression level
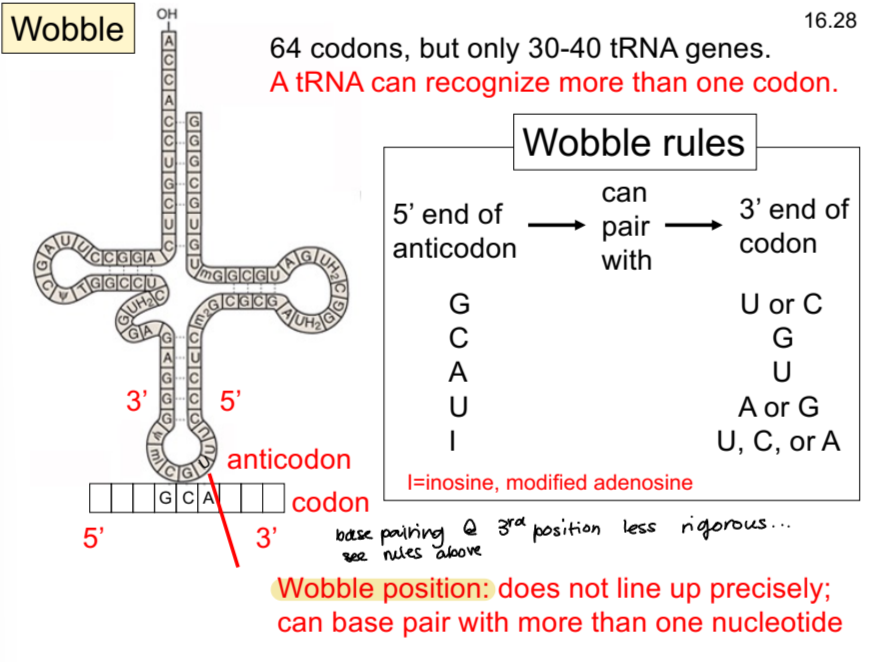
wobble position
bases don’t line up precisely in the 3rd position. . .one nucleotide can base pair with more than one other one
by convention, which strand in dsDNA is coding and which is template?
top —> coding, sense
bottom —> template, anti-sense
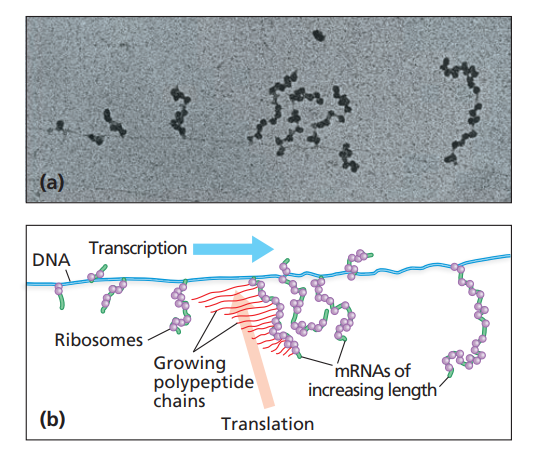
polyribosomes
busy translational complexes containing multiple ribosomes that are each actively translating the same mRNA
synonymous codons
codons that specify the same amino acid