Ch. 10 - Biosynthesis of Nucleic Acids-Replication
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
28 Terms
challenges in DNA replication
continuous unwinding & separation of the 2 DNA strands
must protect unwound portions of DNA from being attacked by nucleases
synthesis of DNA from 5’ to 3’
guard against errors—make sure right bases are added
how long does bacteria take to replicate?
20 mins
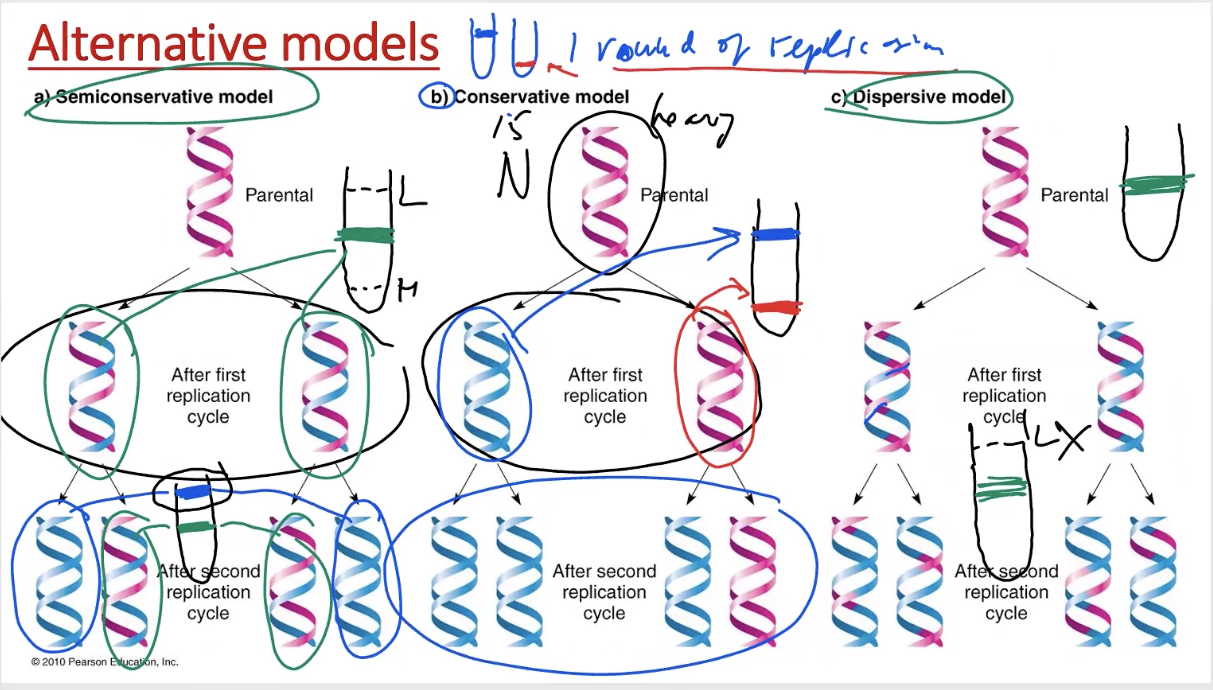
3 ways DNA can be replicated
semiconservative
conservative
dispersive model
semiconservative model
each daughter strand has 1 template strand & 1 newly synthesized one
hybrids make 1 fully new & 1 that’s half-half
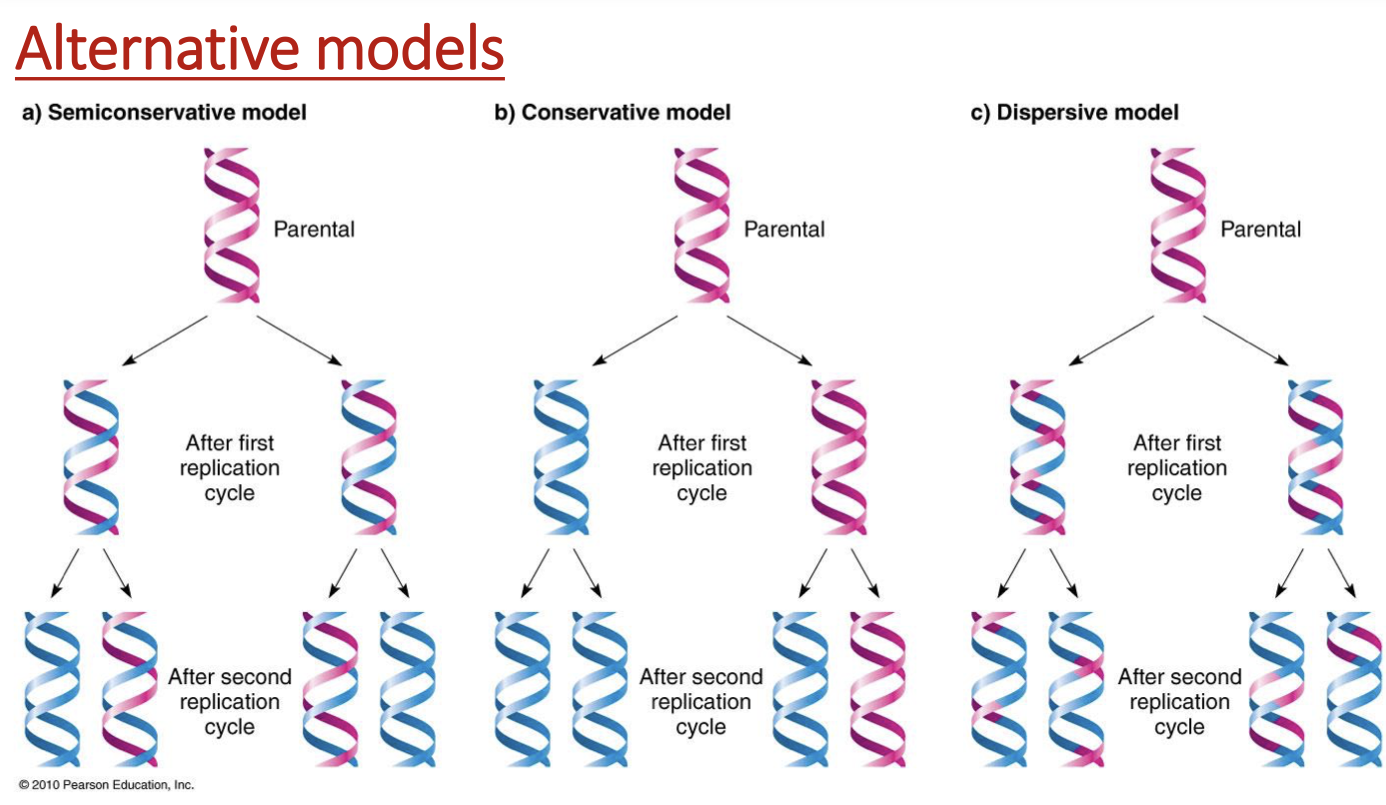
conservative model
1 daughter fully parental, 1 daughter fully newly synthesized
only helices that contain fully parental strands make both
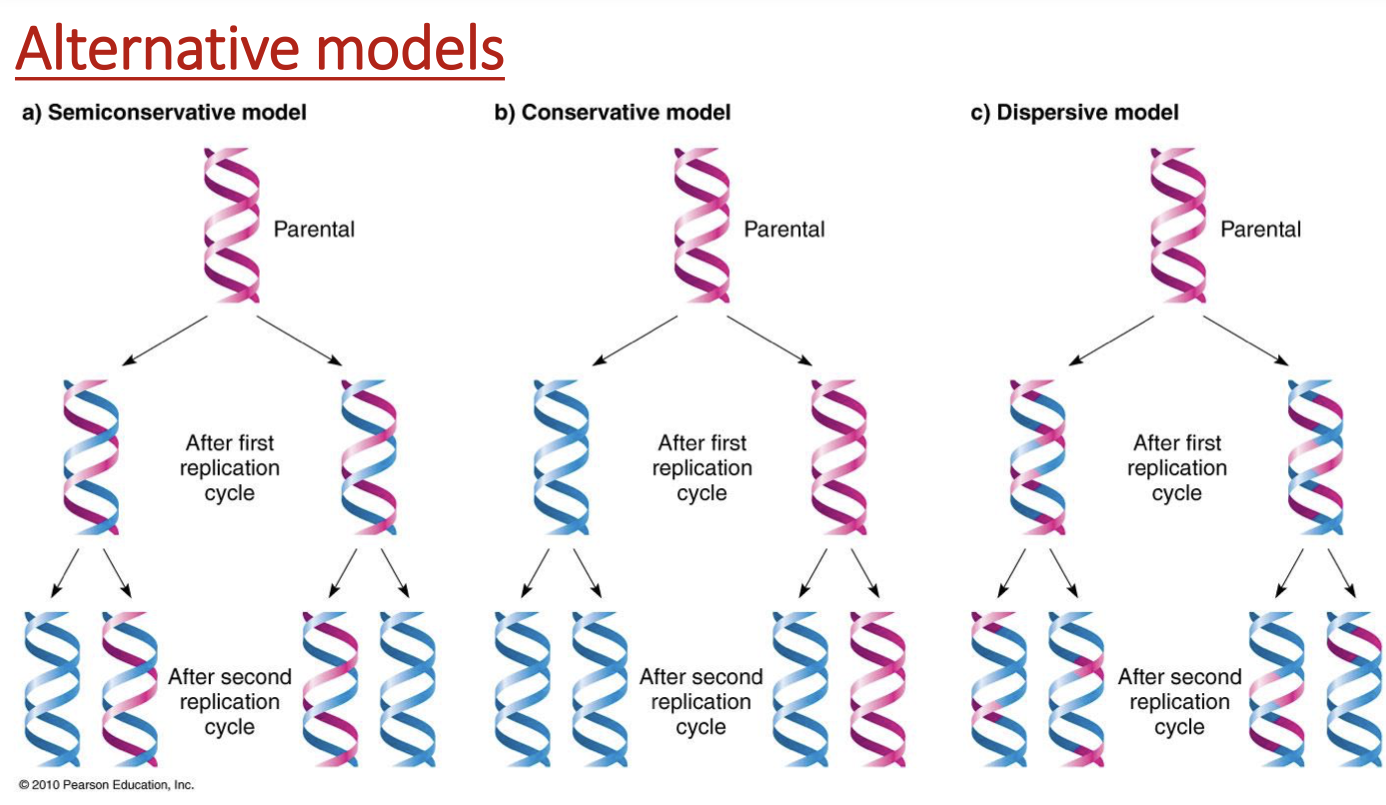
dispersive model
each daughter helix has some parental, some newly synthesized
gradually less & less parental pieces of DNA
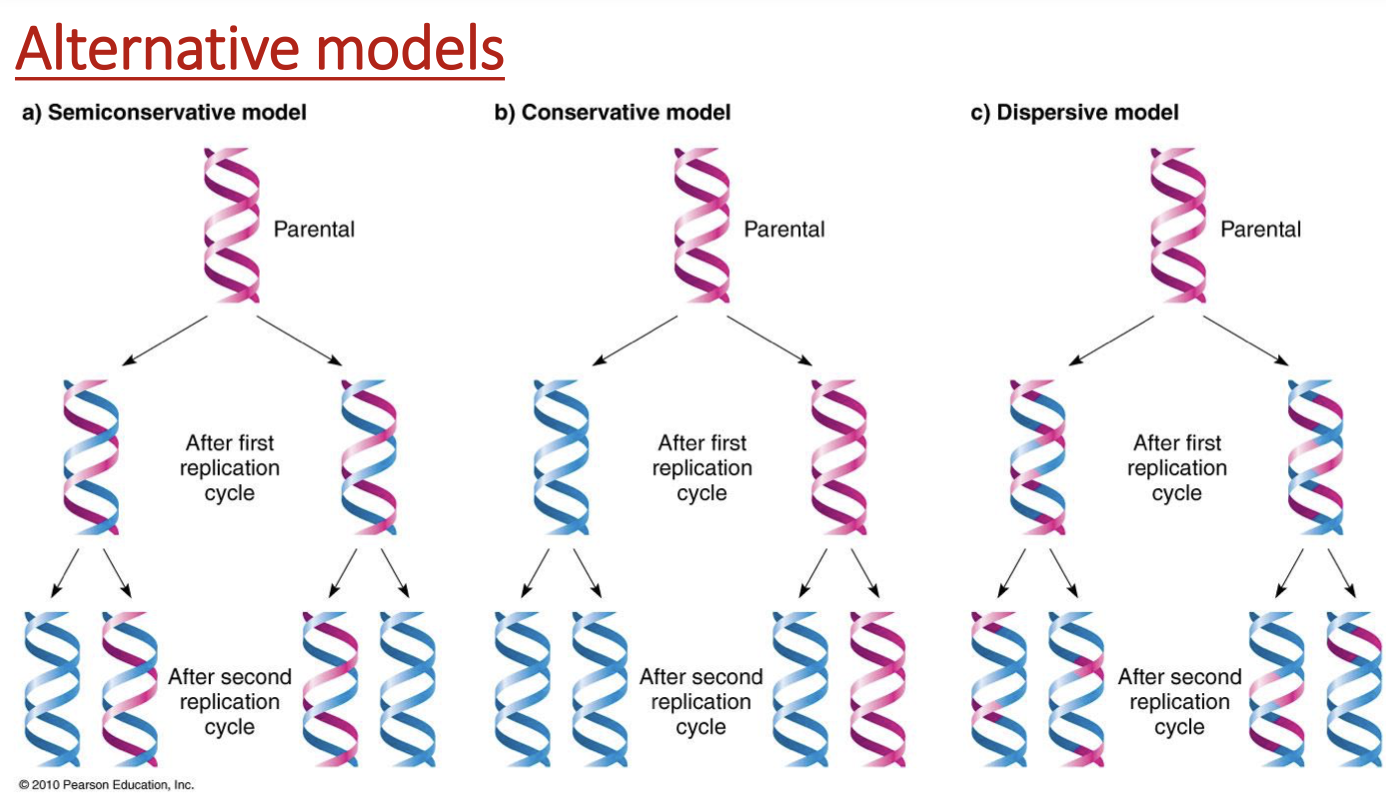
experiment for semiconservative replication
parent DNA labelled w/ N15 (heavy)
first replication in medium N14 (light)
2nd replication in N14
density-gradient centrifugation
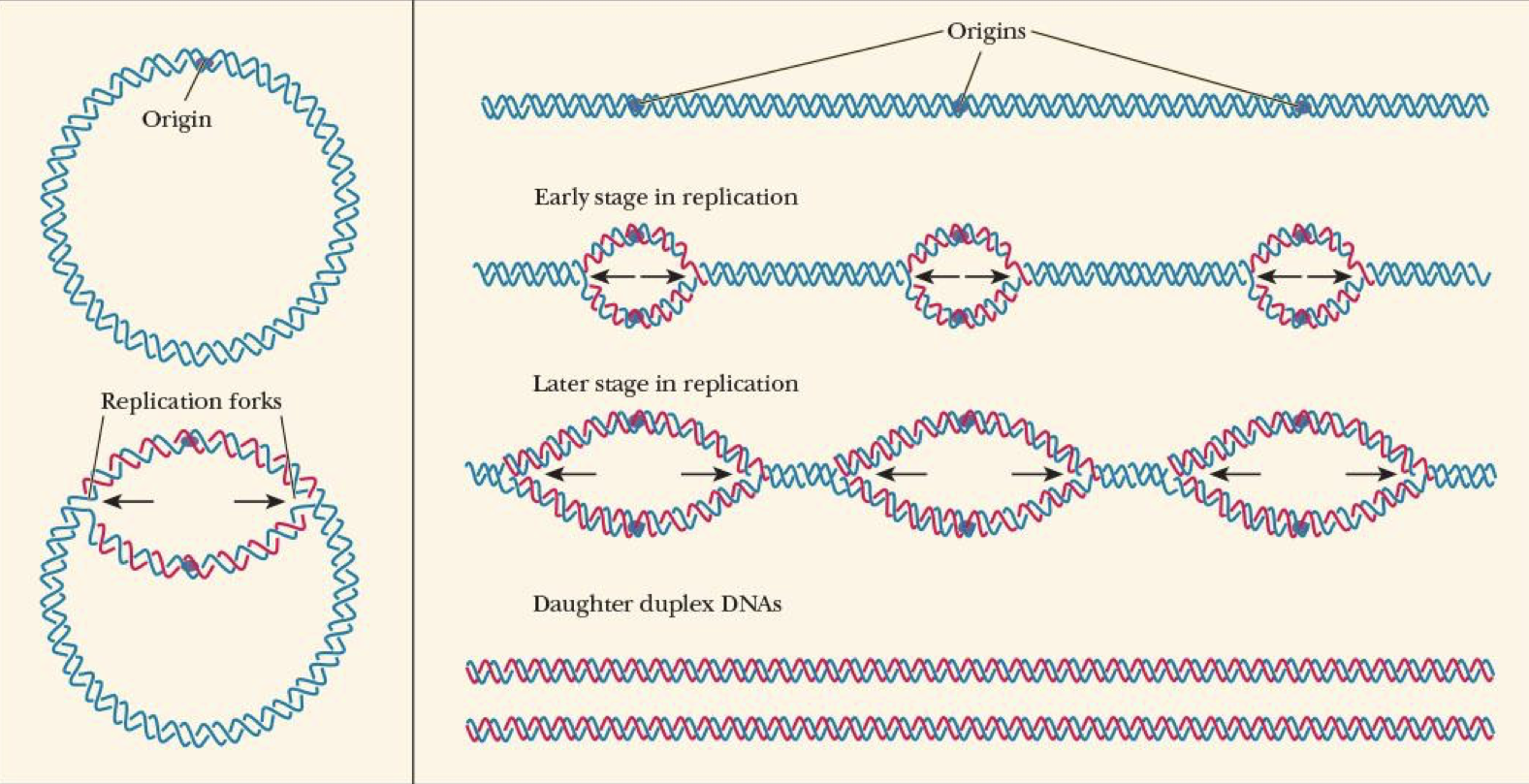
origin of replication
where DNA double helix unwinds
replication forks (looks like 𝛳)
points where new polynucleotides are formed
usually 2 bc DNA replication is bidirectional in most organisms
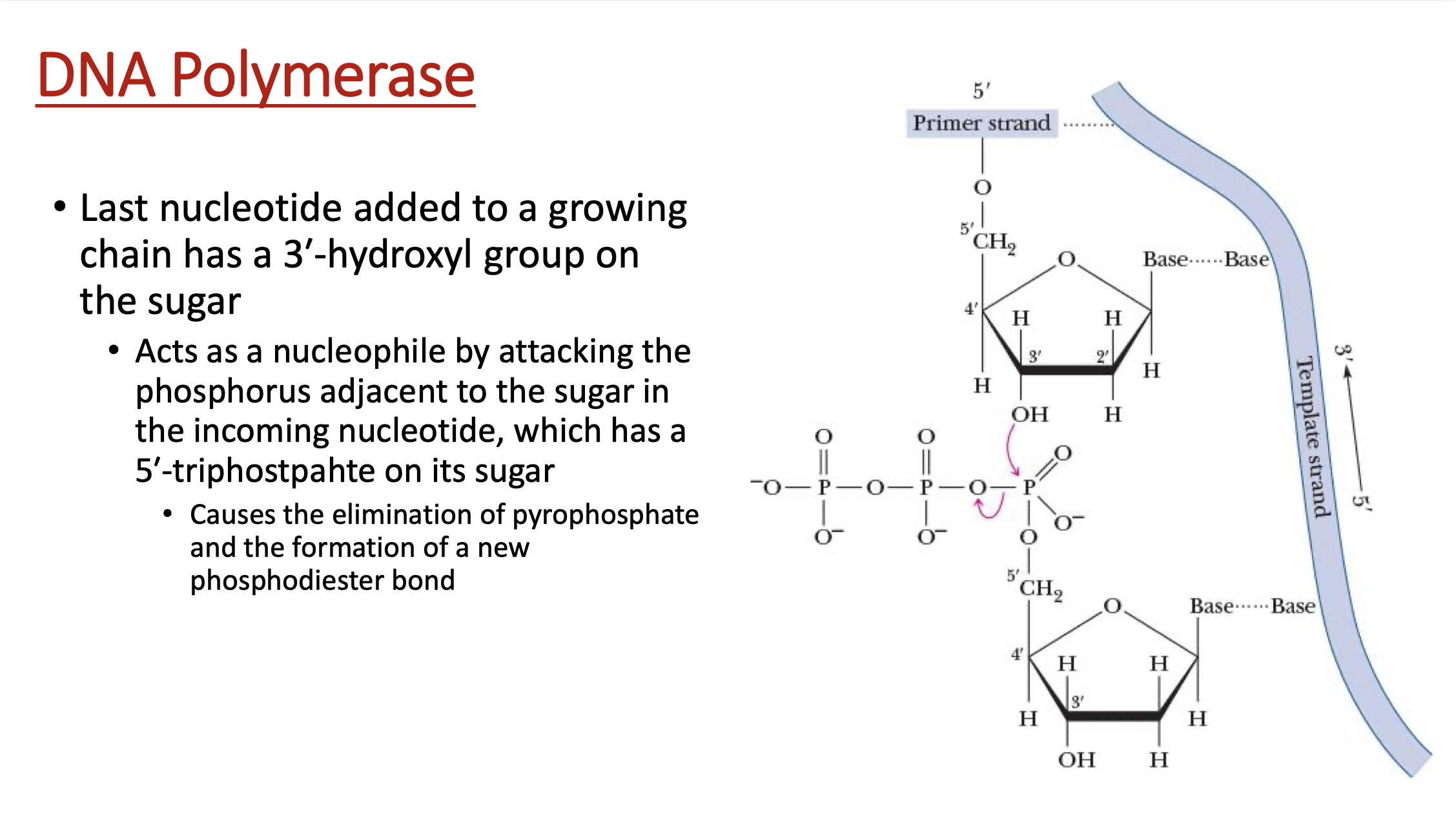
explain how DNA polymerase works
dNTP gets attacked nucleophillically by OH group on 3’ carbon of sugar
PPi leaves & new phosphodiester bond is formed, connecting to the following sugar’s 5’
leading strand
synthesized continuously from 5’ to 3’ at replication fork on 3’ to 5’ exposed template strand
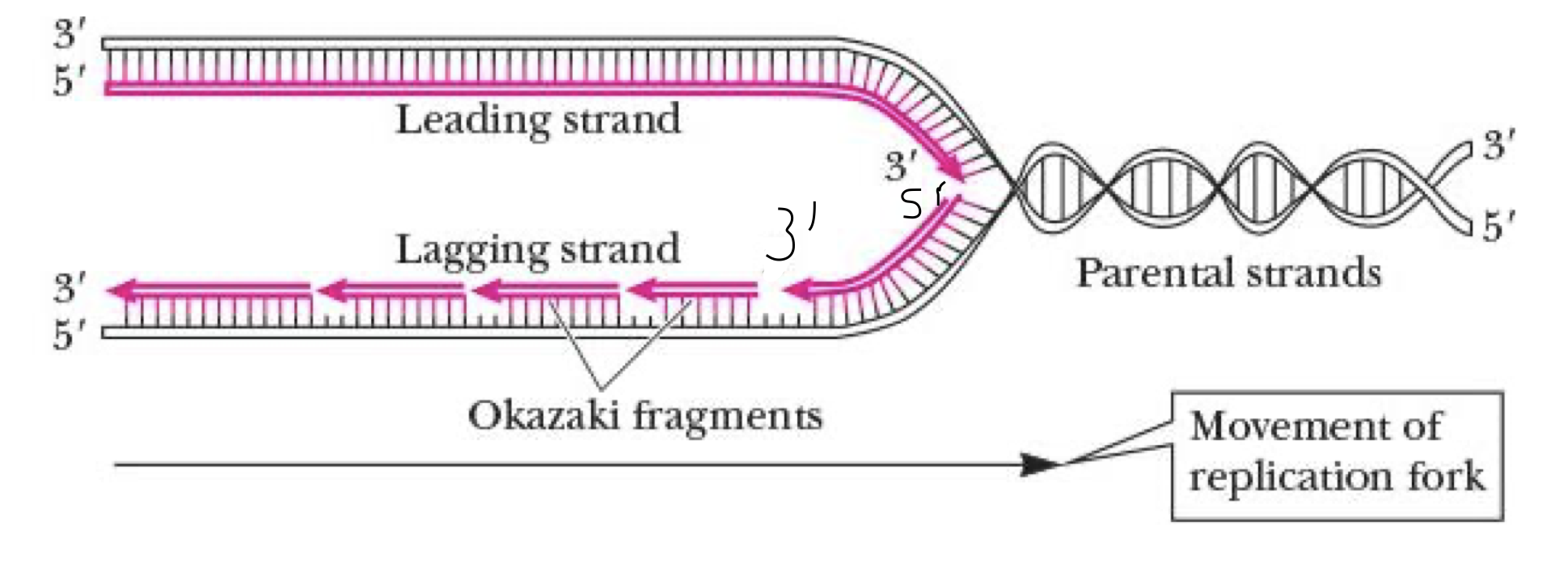
lagging strand
synthesized discontinuously in Okazaki fragments
fragments linked tgt by DNA ligase
still needs to face same way as the leading strand so it loops a bit
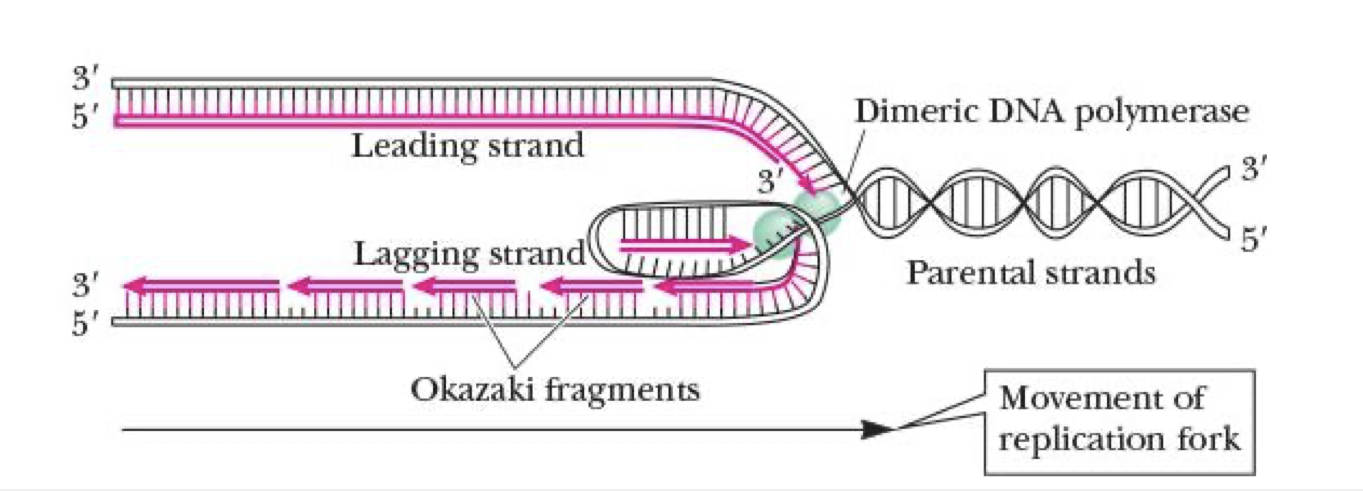
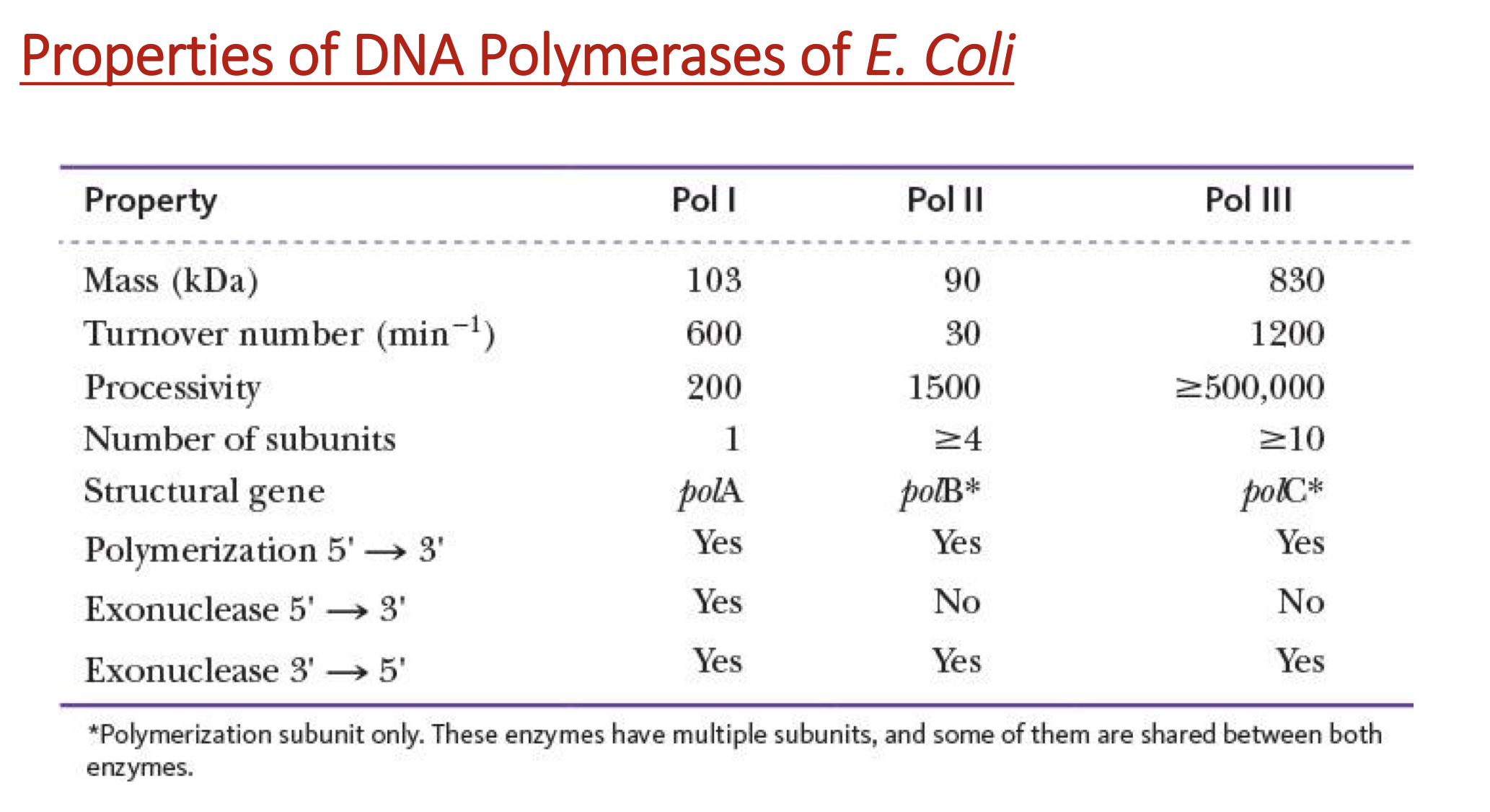
how many DNA polymerases in E. coli?
5
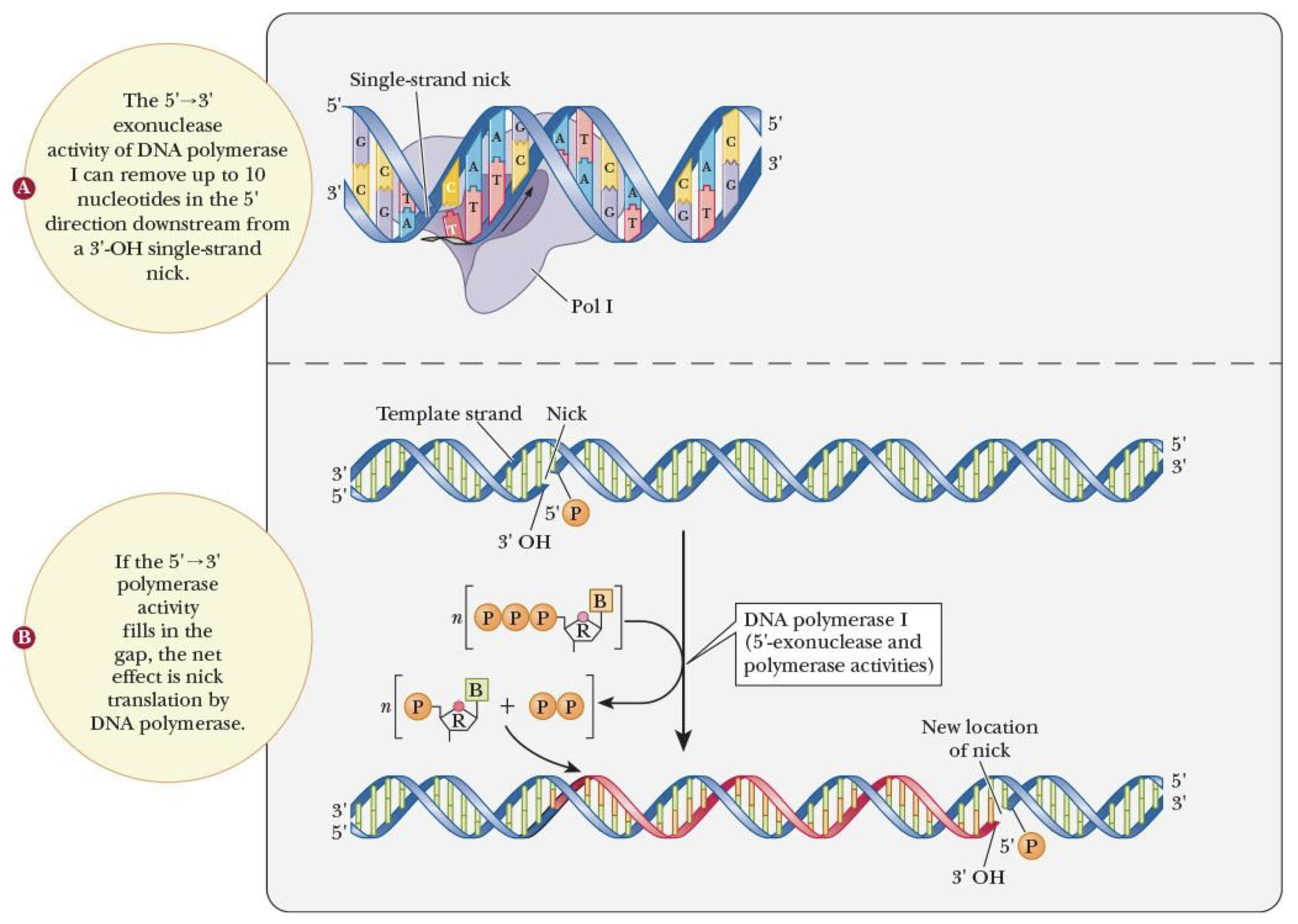
Pol I
single polypeptide chain
repairing & patching DNA
Pol II & III
multi subunit proteins that share some common subunits
Pol III → polymerization of the new DNA strand bc turnover # (per min) > 500,000
Pol II, IV, V
repairing enzymes
processivity
# of nucleotides incorporated before the polymerase dissociates from the template
exonuclease
enzyme that remove nucleotides from an end of RNA to correct errors made
DNA polymerase can’t catalyze de novo synthesis (from scratch), what do they need?
primer (10-20 bases)
needs the 4 ribonucleoside triphosphates
DNTPs
DNA template
Mg2+
replisomes
complex of:
DNA polymerase
RNA primer
primase
helicase
at replication fork
DNA gyrase (class II topoisomerase)
fights positive supercoils
energy from ATP
helicase
helix-destabilizing protein
promotes unwinding DNA by binding at replication fork
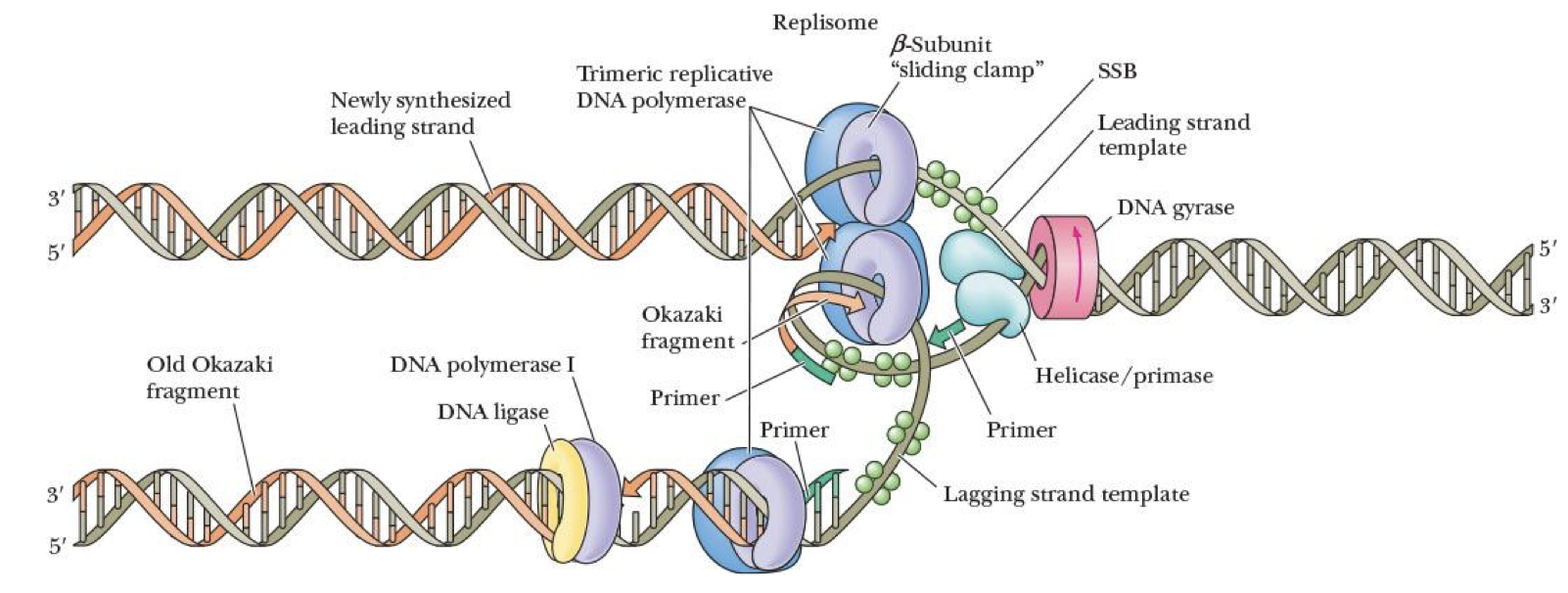
SSB (single-strand binding protein)
stabilizes single-stranded regions by binding to them
protects them from being destroyed bc our bodies recognize single-stranded DNA as viruses
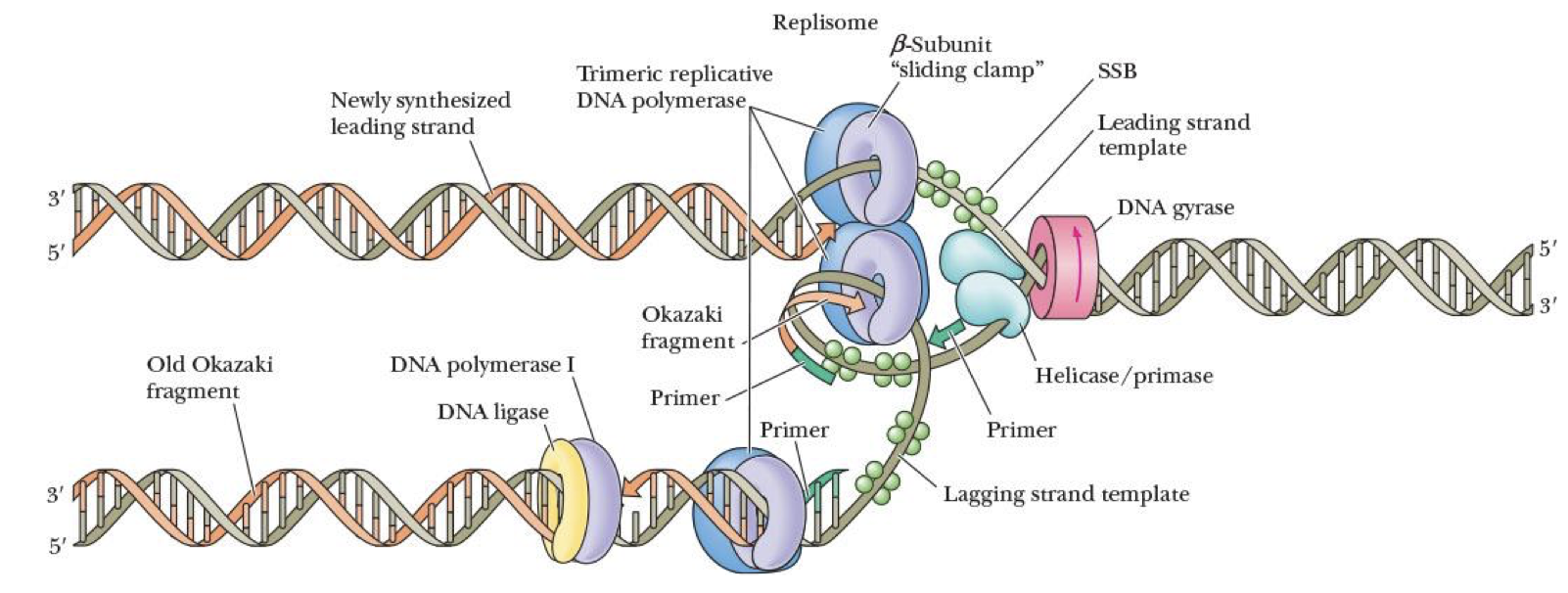
primase
enzyme that makes short section of RNA to act as primer for DNA synthesis
primosome
complex at replication fork consisting of RNA primer, primase, & helicase
frequency of mutations
once every 109–1010 (1–10 billion) base pairs
proofreading
removal of incorrect nucleotides right after they’re added to growing DNA
reduces errors from once every 104-105 base pairs to 109–1010
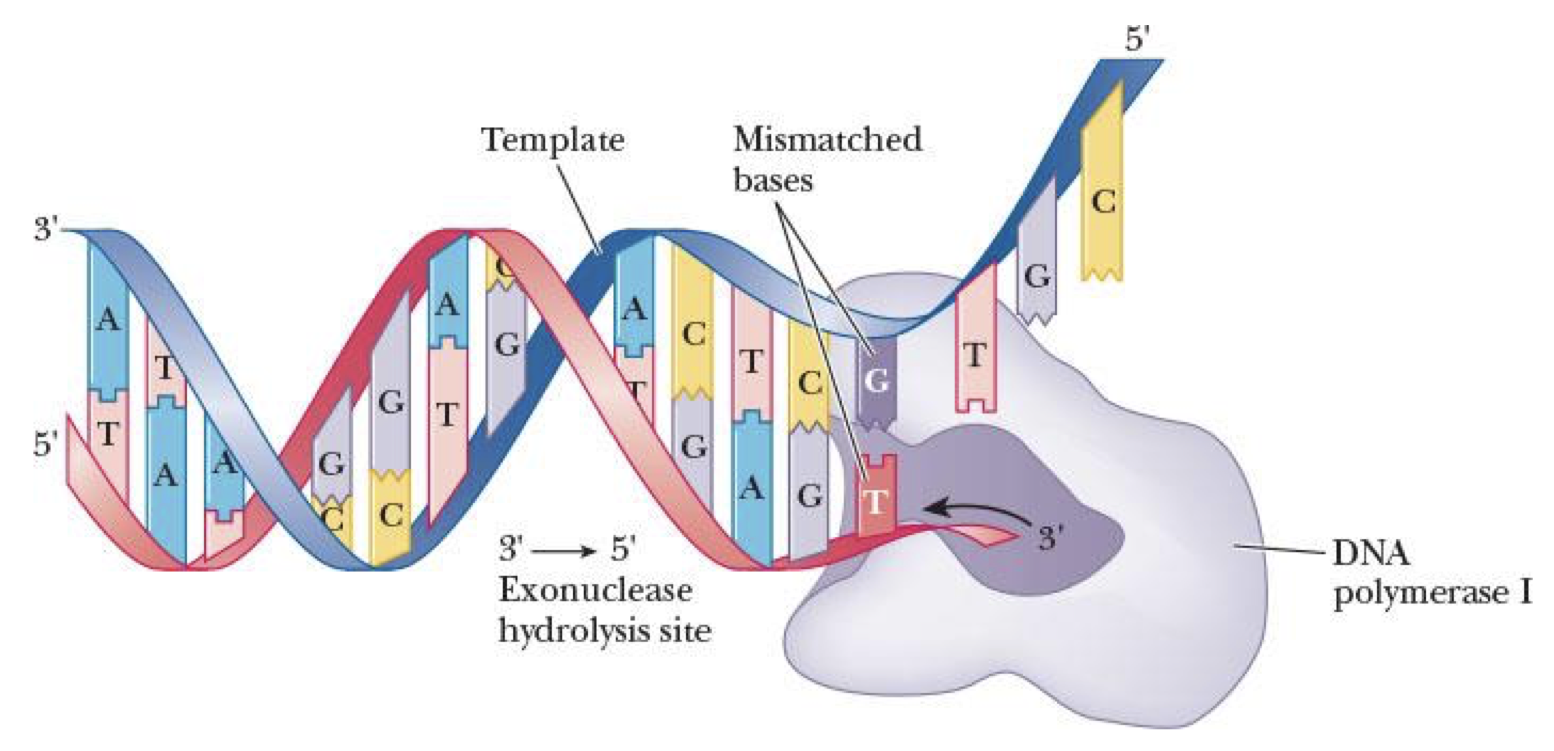
nick translation (mainly for reference only)
removal of RNA primers or DNA mistakes by Pol I using 5’–3’ exonuclease activity & filling in behind it w/ its polymerase activity