BIN300 -W3 Design of QTL mapping experiments
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
linkage marker -QTL gene
many similariteis, differentL WTL genotyp is not observed, try to infer QTL genotype from phenotype
old markers
protein polymorphisms, might have an effect on trait
new markers: RFLP, microsatellitles, SNP
Unlikely to have effect on trait
population wide LD mapping (additive effect a)
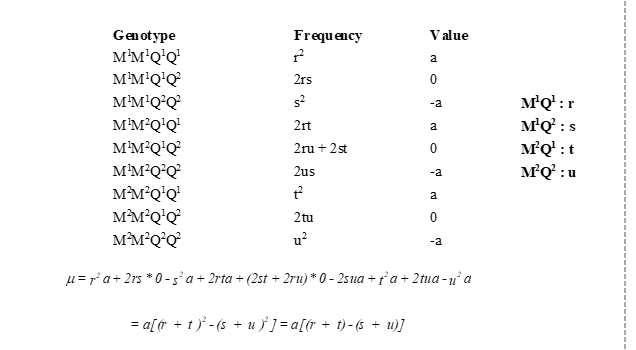
Linkage disequilibrium (LD)
alleles at locus 1 are non-randomly associated with alleles at locus 2
D is difference between observed haplotypes
R2 standardized measure
D=0, are in linkage equilirbium, not 0 in LD
R2 = 0, no LD, R2 is 1, perfect LD, one ALLELE can predict other
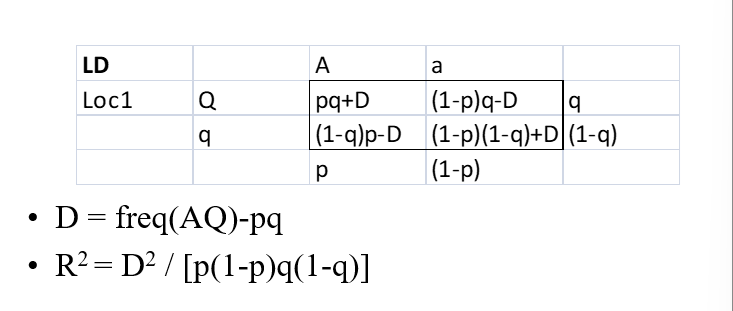
WTL genotypes and values
QQ: m+a (m=mean = effect of all other QTL)
Qq: m+
no dominance, d=0, complete dominance d=a
qq: m-a
d
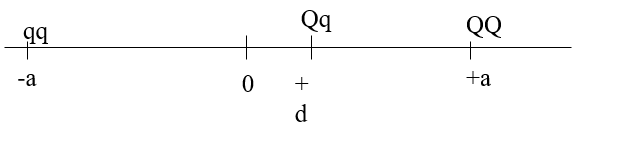
Means of marker genotypes
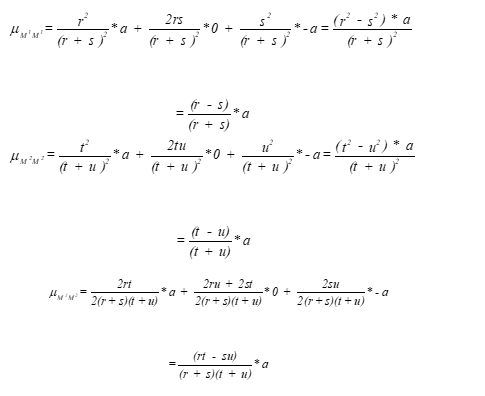
difference between homozygotes
D=r*u-s*t
D=0 → no effect of genotypes
D=1 → full effect of marker genotypes
Significant difference: could be gene or linked marker
marker has effect if D does not equal 0
marker = gene
recent mutation, random drift, selection, or migration
recombinaiton reduces D
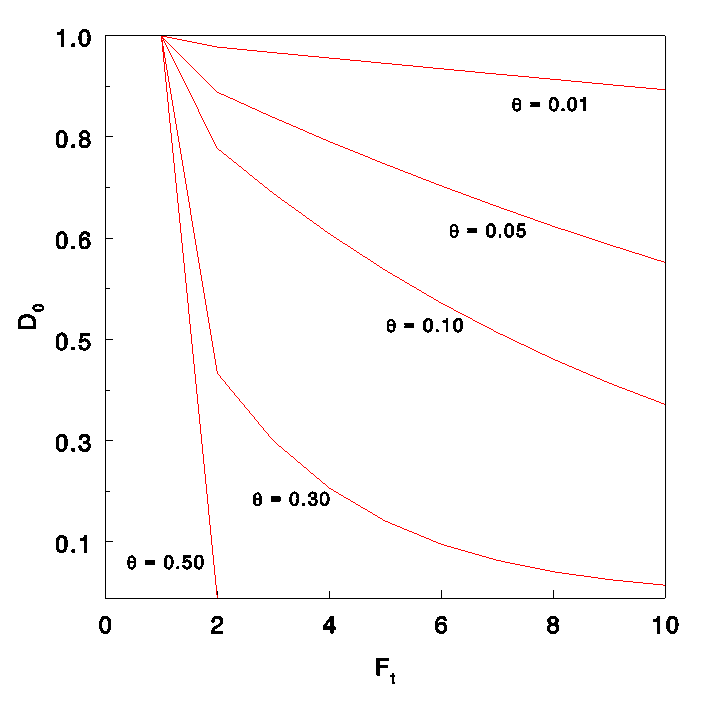
Dt=D0 (1-Ɵ)^t
Ɵ small Dt (still) large
need dense marker map
D0 large, crosses between lines
QTL dtection in crosses between inbred lines
F1 animals are heterozygous, complete LD, QTL and marker freqs = 0.5, linakge phase is as in parents
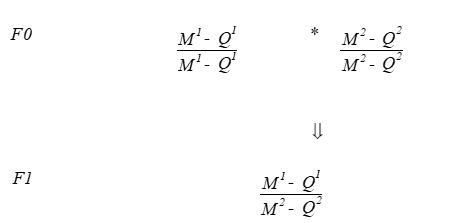
The back-cross design
M1Q1/M2Q2 * M2Q2/M2Q2

The F2- design
M1A1/M2Q2 * M1Q1/M2Q2
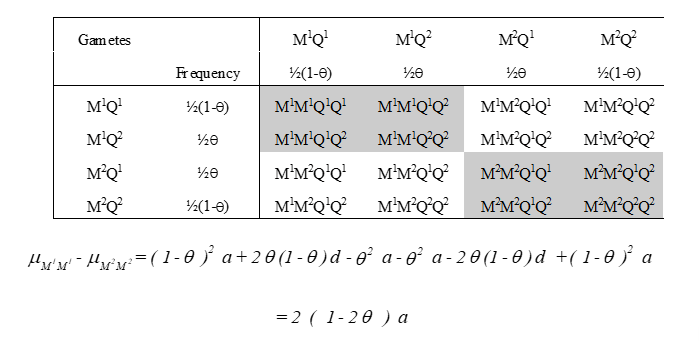
M1M1-M2M2 = 2(1-Ɵ)A
size of QTL effect: a
distance from marker till QTL
a and Ɵ cannot be disentangled, although maximum likelihood can, but power is lown
no fixation at QTL/marke
marker fixed/QTL not

marker and QTL not fixed, in terms of D
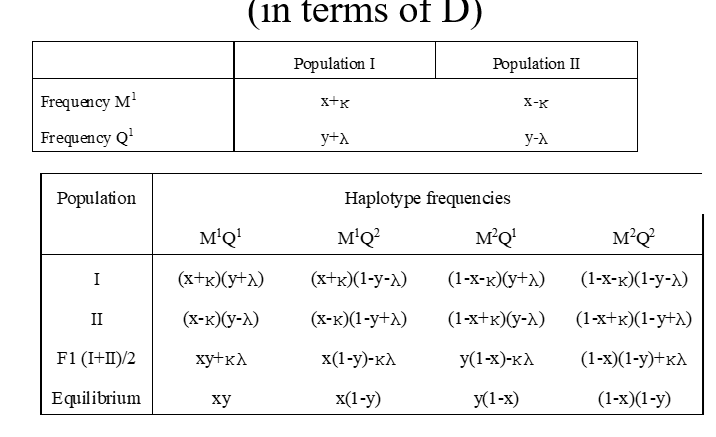
reduction of LD
coupling phase: M1Q1/M2Q2
M1Q2 with freq ½ Ɵ, i.e. reduction of M1Q1 haplo of 1/2Ɵ
repulsion phase: M1Q2/M2Q1
M1Q1 with freq 1/2Ɵ, i.e. increase of M1Q1 haplo of ½ Ɵ
chan
change in freq M1Q1
1/2Ɵ [freq{repulsion)-freq(coupling)]=
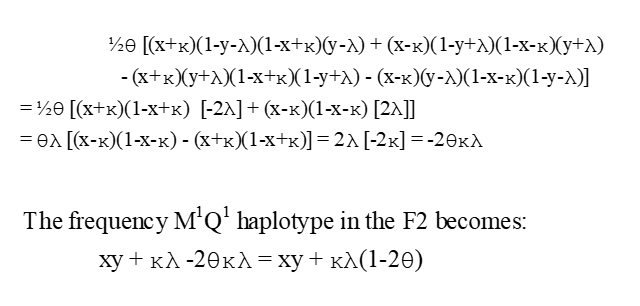
freq M1Q1 in F3 and FTS
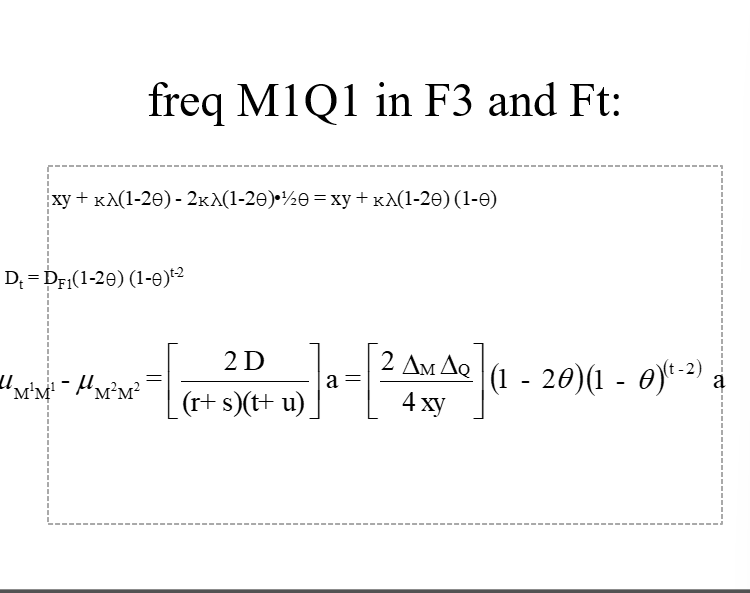
LD as a function of time and Ɵ
crosses of inbred lines
useful in plants and lab animals
in animals approximate by wildboar*largewhite
useful for detecting QTL
is QTL also segregating within population?
is +allele always present in largewhite?