Bacterial Genes and Genetics: chromosomes, plasmids and gene transfer
1/32
Earn XP
Description and Tags
All 3 lectures
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
33 Terms
Describe E.coli
First choice for basic bacterial genetics. When is lysed (make holes in) for electron microscopy the DNA spews out (it is 1500x longer than the cell).
The nucleoid is NOT a nucleus, true or false?
True.
Define nucleoid
Not nucleus, no membrane. Contains all or most genetic info.
Define histones
Keep chromosomes of cells in ordered + supercoiled way.
Define supercoiling
Under or overwinding of bacterial DNA which impacts levels of chromosome structure.
How is supercoiled state of a nucleoid change?
Enzymes (topoisomerases) make a double-stranded break in the circle, passes another part of DNA through the break then reseals it.
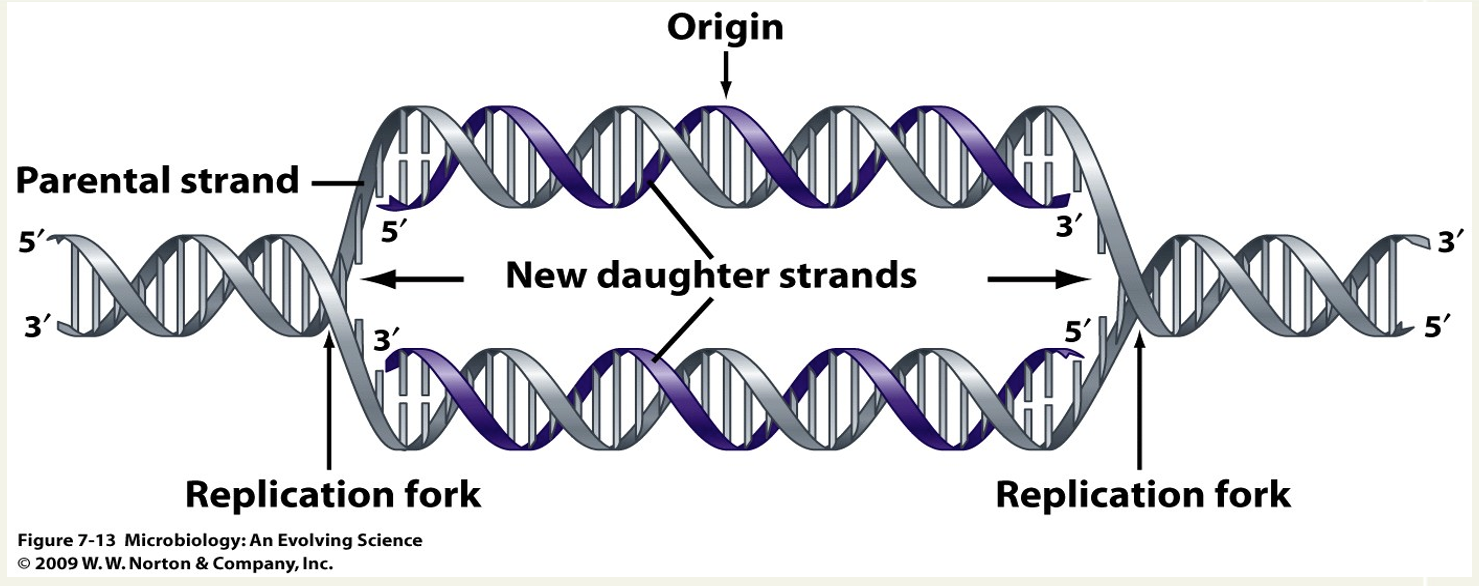
Define semiconservative replication
At the replication fork, the DNA synthesis machine separates the 2 strands while extending the new growing strands. End up with one old strand + one new/replicated strand.
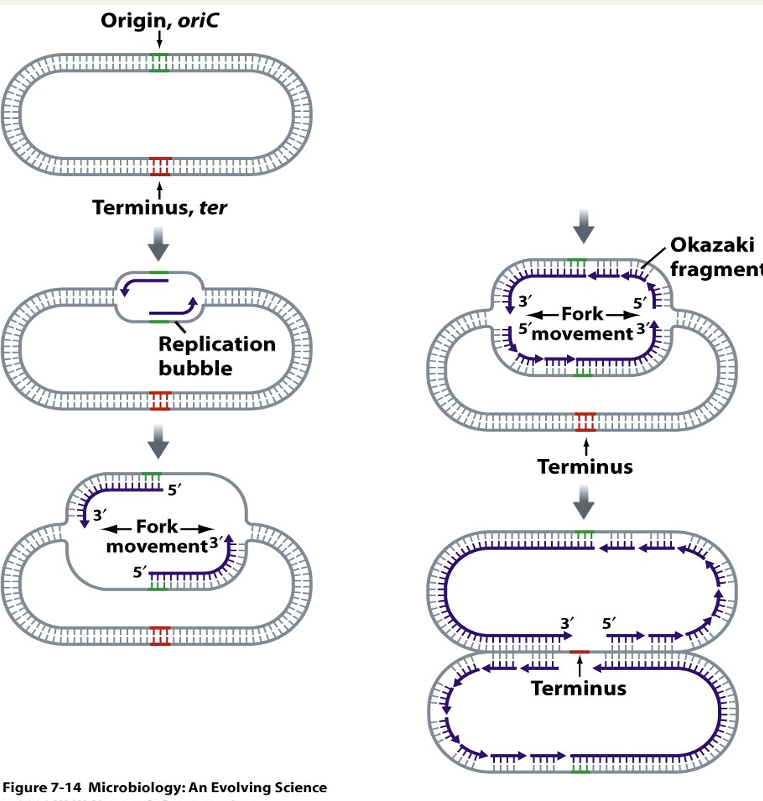
Describe replication bubble
Point of origin where 2 strands are opened so there is enough space for DNA polymerase to do its job so DNA can be copied. Is a double job as some machinery works clockwise + some anticlockwise.
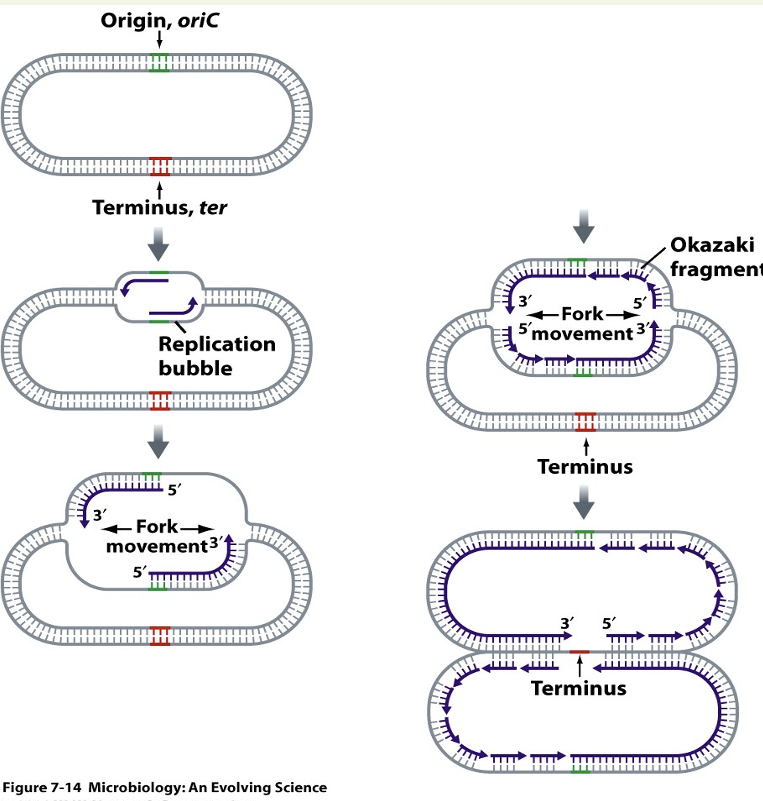
Describe replication forks
2 are produced. Point on DNA that is opened up + started to be copied, 4 strands open (one copy of top + one of bottom). DNA polymerase can only synthesise from 5’ to 3’: works for bottom strand but not top as complementary = Okazaki fragments.
Define Okazaki fragments
Small pieces of DNA. DNA polymerase can only synthesise from 5’ to 3’: works for bottom strand but not top as complementary = when fork opens it puts small amount of sequence that is 3’ so DNA polymerase can work then adds another small amount etc = small gap so ligases join small pieces of strands.
How is DNA replication terminated?
Terminator regions for DNA replication on the E.coli chromosome.
There are >8 arrest sites at the terminus.
Replication forks moving clockwise are trapped by terB, C, F, G, J.
Counterclockwise: moving forks are trapped by terA, D, E, H, I.
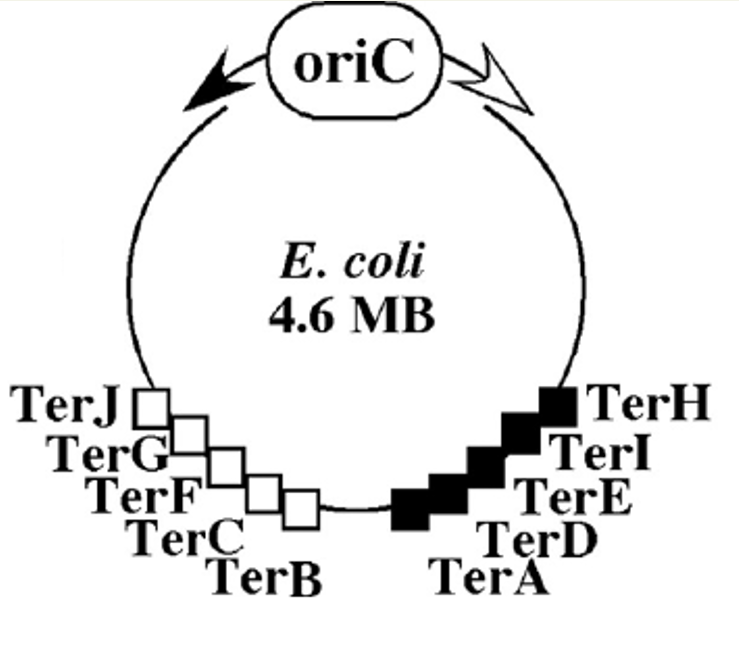
Define terminus
aka ter, sequence that stops rejection.
Describe replication clockwise
Goes very fast then caught by proteins + binding + waiting for DNA polymerase to arrive = bubble + fork destroyed.
Describe replication anticlockwise
Same as clockwise but different proteins involved.
What are the differences between the chromosomes of eukaryotes and prokaryotes?
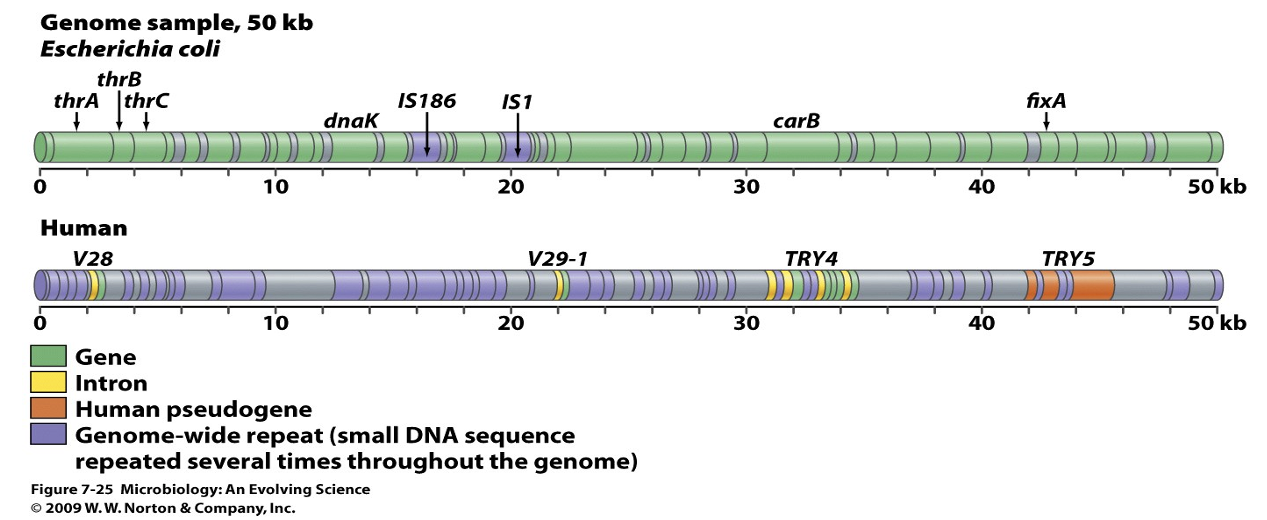
Bacterial chromosome is tightly packed with coding genes (green) with few mobile elements + a little unused sequence (purple). Human chromosome has 95% non-coding sequences (purple), coding regions are separated by introns (yellow), ancient gene duplications have decayed to pseudogenes (orange).
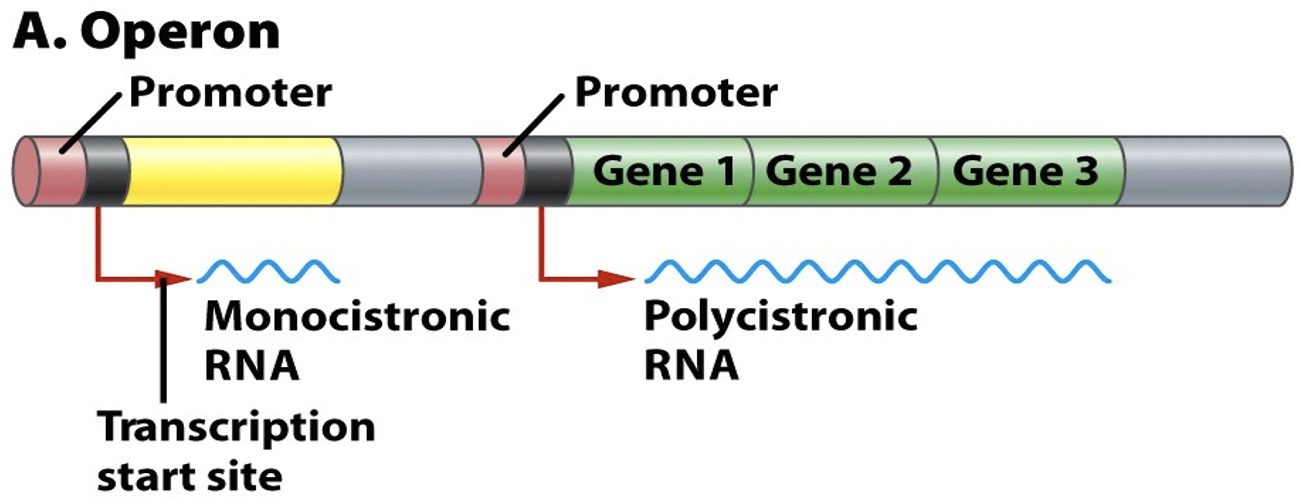
Describe operons
One single gene (yellow) makes a monocistronic message (for one protein). An operons of three genes (green) makes a polycistronic message.
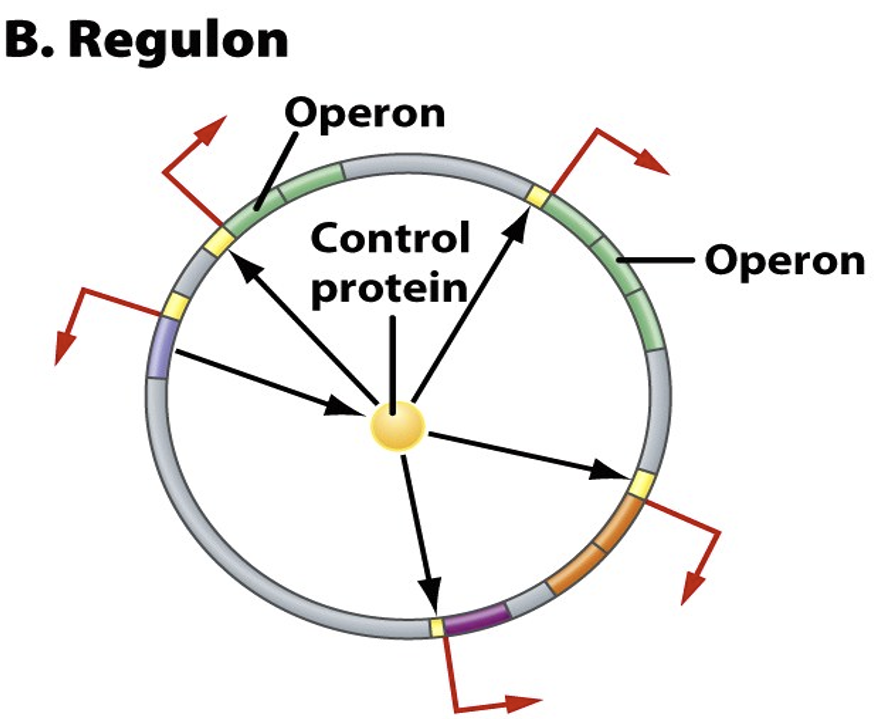
Define promoter
Sequence recognised by mRNA so message is translated to protein. Is present if a gene needs induced. Protein attaches to top of promoter if protein is not needed = prevents it from being transcribed.
Describe regulon
In chromosomes, formed of operons + genes. Controlled by a common regulatory protein.
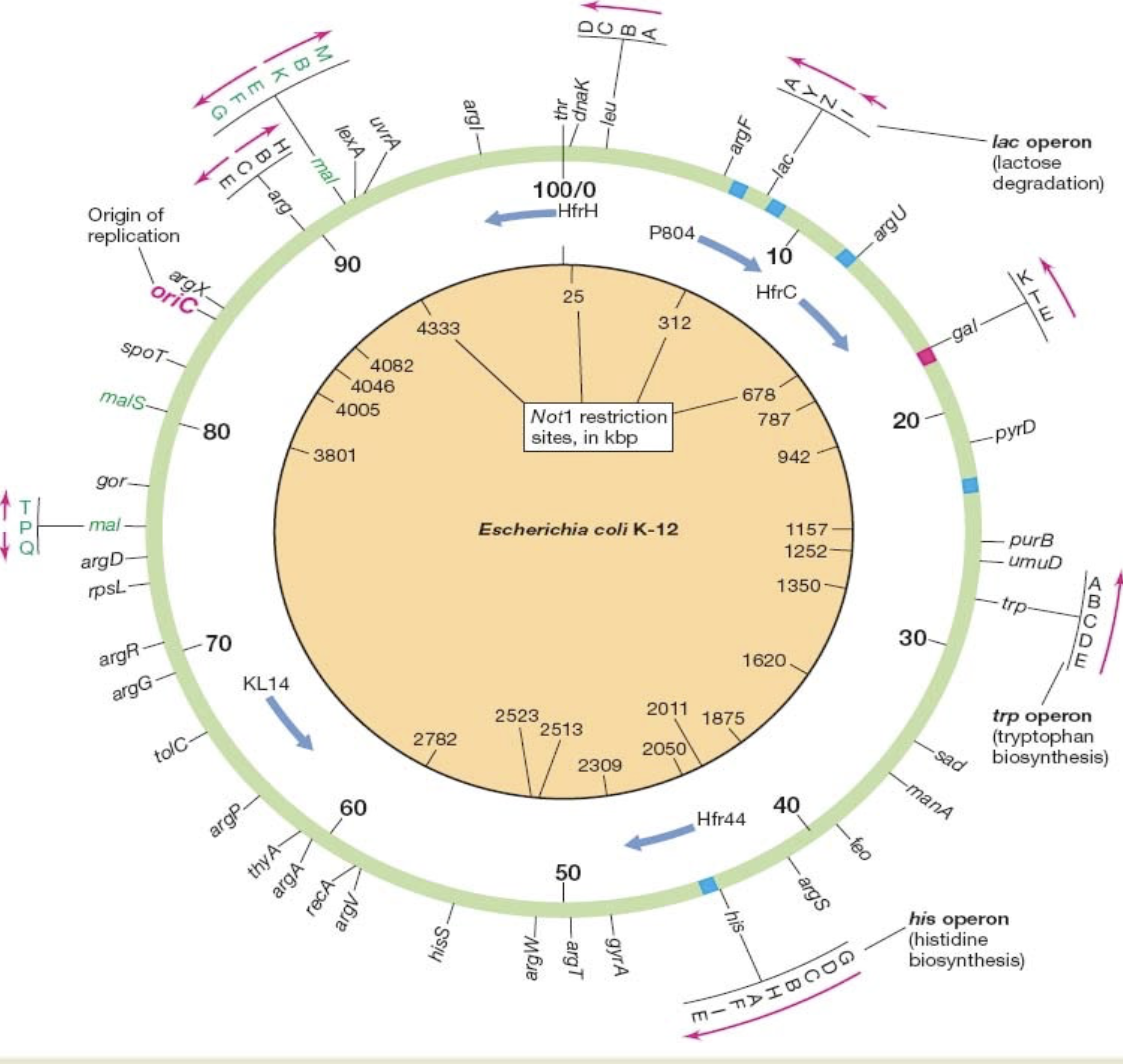
Describe the chromosome of E.coli
Circular. 4,288 genes. Some genes + operons are shown outside. Order of transcription can be clockwise OR counterclockwise. Replication proceeds in BOTH directions from the origin at 84.3 min. Restriction sites shown inside are for Not1. Some Hfr origins are also shown.
Define plasmid
A genetic structure in a cell that can replicate independently of the chromosomes, typically a small circular DNA strand in the cytoplasm of a bacterium.
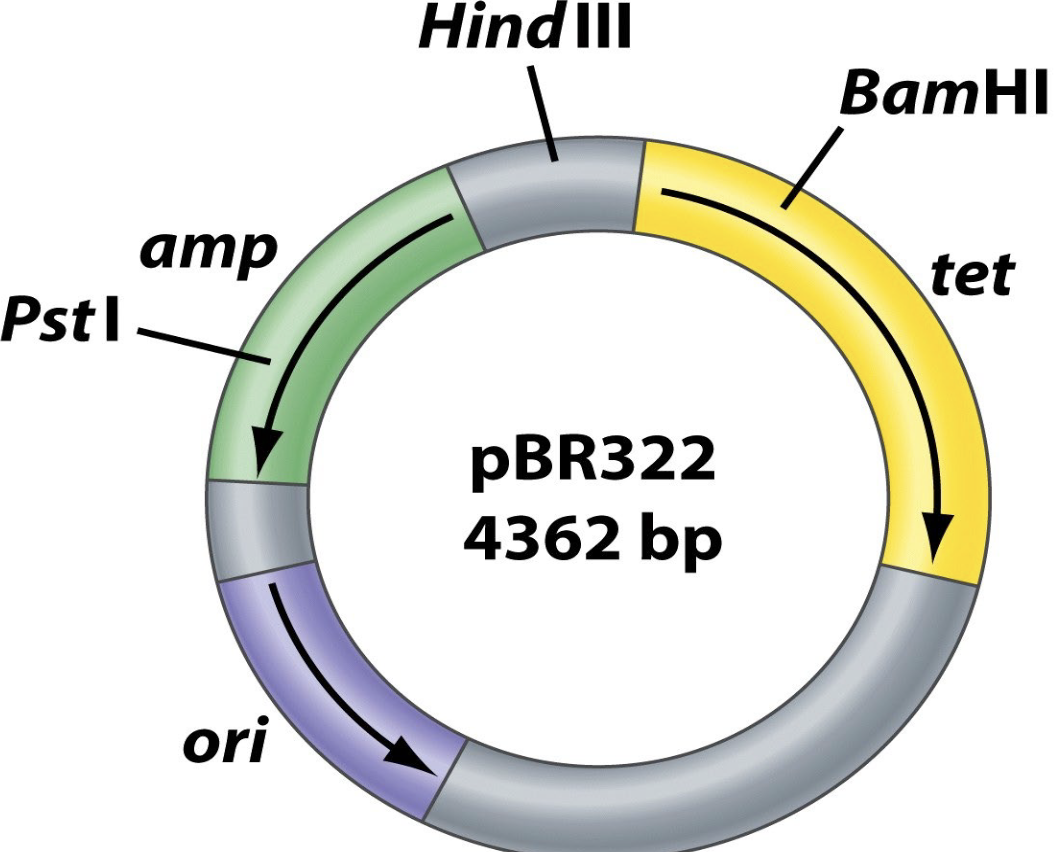
Describe the common cloning vector or plasmid pBR322
Is widely used in molecular cloning. Possesses: an origin of replication, 2 antibiotic resistance genes (amp, tet), 3 unique 6-bp restriction sites.
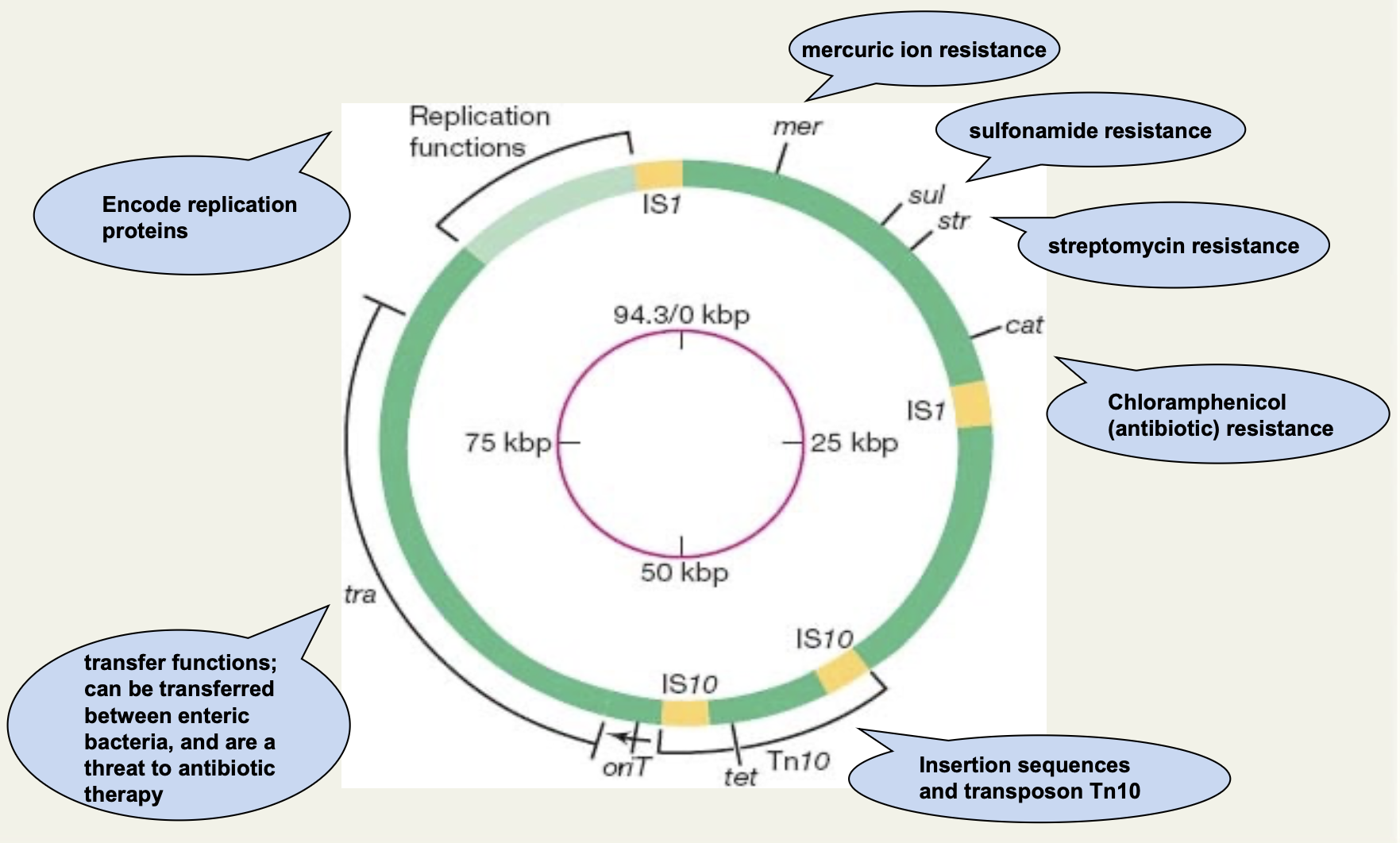
Describe genetic map of the resistance plasmid R100
Encode replication proteins.
Define state features of a mutation
Heritable, rare, can be good bad or neutral, usually ‘small’.
State genotype nomenclature rules
Gene name is given as 3 or 4 letters.
All except the 4th in lower case.
Always italicised.
A wild-type gene is written cydA^+ (for example).
A mutant gene is written cydA (no superscript ‘minus’)
If there are several alleles, write cydA1, cydA2 etc.
State phenotype nomenclature rules
A plus or minus is used to designate presence or absence of a property. Not in italics. Only first letter in capitals.
State encoded protein nomenclature rules
No superscript, not in italics, only first letter is in capitals.
The genetic code is degenerate, what does this mean?
More than one codon can specify an a.a.
What can single codon changes result in?
Either silent, missense or nonsense mutations.
What can specific mutagens cause?
Various effects = can be used to induce specific mutations.
State 3 ways bacteria can get genetic material
Transformation (up-take of naked DNA), transduction (phage-mediated), conjungation (plasmid-mediated).
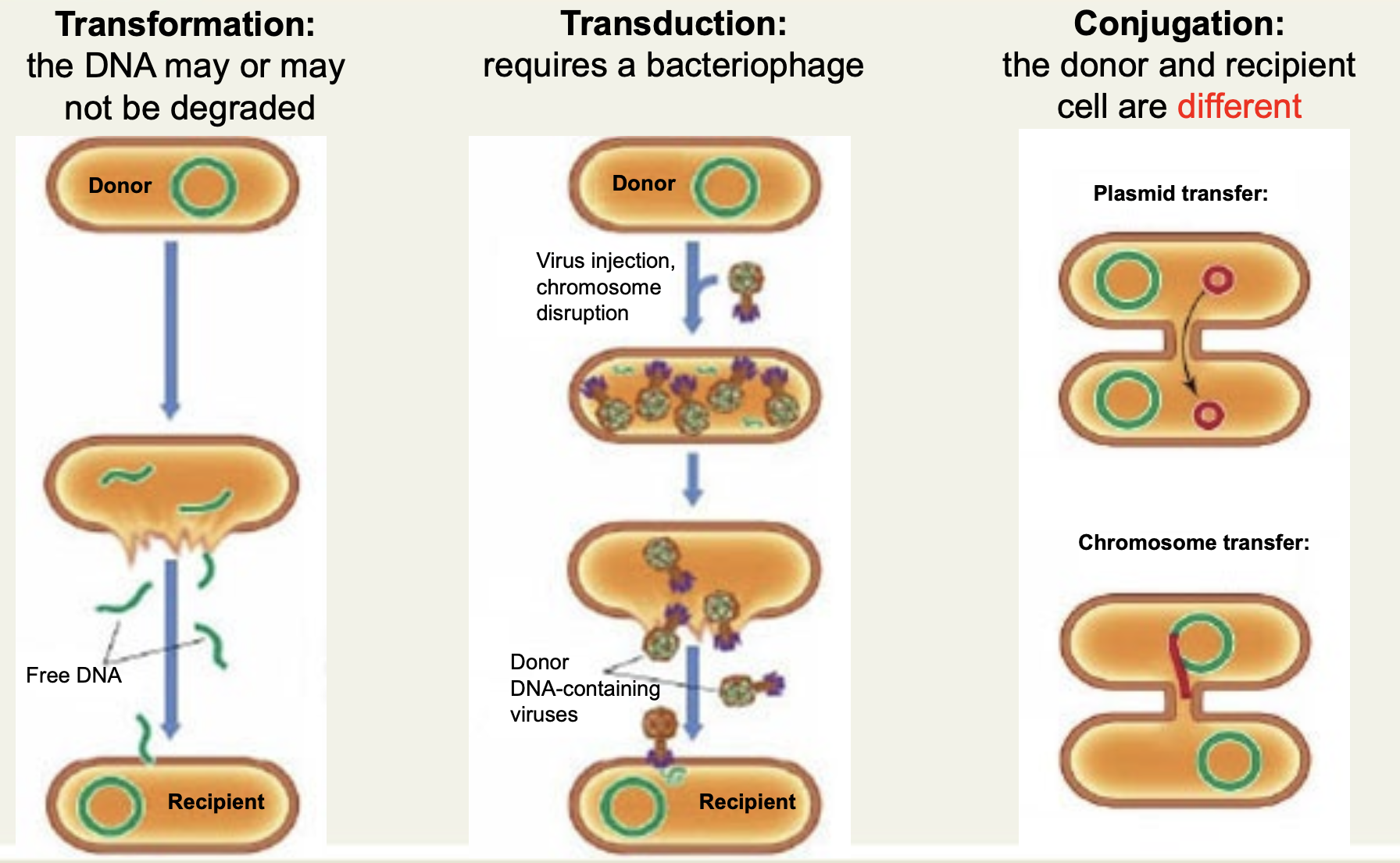
State the mechanism of transformation
DNA binds via protein.
ONE strand enters using the DNA translocase, the second is degraded.
The internalised single strand is bound by RecA protein.
As RecA has more than one DNA binding site, it can hold the ssDNA + dsDNA together.
The RecA-ssDNA complex stretches the dsDNA to increase complentarity recognition (conformational proofreading).
Branch migration + homologous recombination follows.
Transforming DNA doesn't need to be a plasmid but often is.

Define naturally competent
Can take DNA naturally.
Describe the mechanism of generalised transduction