MCB104 - Lecture 13 - Epigenetics and Genome Sequencing
1/64
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
65 Terms
What is epigenetics?
“Stuff on top of genetics”
DNA sequence in genes is not the only carrier of genetic information
Epigenetic phenomena: heritable self perpetuating changes in gene expression not caused by base pair sequence changes
Usually involves modified cytosine residues, modified histone tails in chromatin, small RNAs
Epigenetic factors that can determine whether a gene is “on” or “off” can change

How can 2 mice have the same genotype for coat color?
Epigenetics can change phenotype
Yellow mouse expresses yellow coat color but grey mouse has coat color gene silenced form epigenetic modifications
Epigenetic information can therefore determine if gene is “on” or “off”
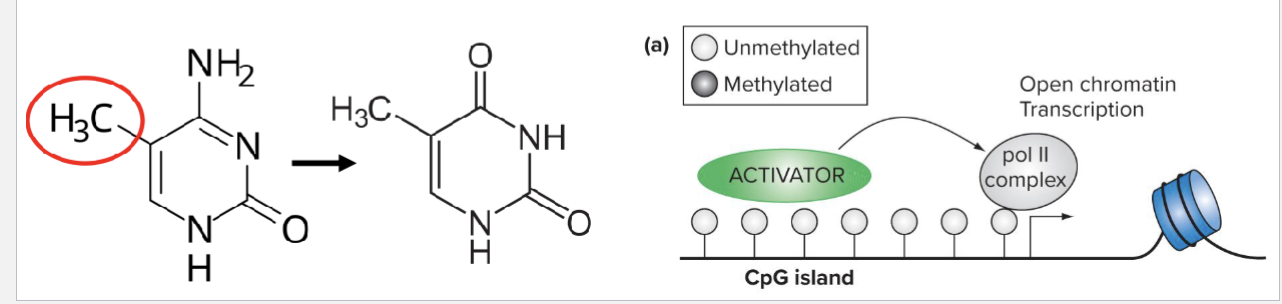
What is DNA methylation?
A methyl group is added to the cytosine base in a 5’ CpG 3’ dinucleotide by DNA methyl transferases (DNMTs)
DNA methylation maintained through cell division
Special DNMT at the replication fork methylates the newly synthesized DNA strand
What is the ROLE of DNA methylation?
CpG methylation turns OFF gene expression
Methyl-CpG-binding proteins (MeCPs) bind to methylated CpG islands and close the chromatin structure
These are transcriptional repressors
What are CpG Islands?
Deamination of methylated Cytosine results in Thymine
This means that frequently methylated CpGs tend to be lost
As active promoters tend to be unmethylated, CpG islands tend to be found in their vicinity
What are Histone Tail Modifications?
Modifying histone tails alter affinity to DNA
Methylation of Lysine decreased negative charge, increasing affinity to DNA
Acetylation of Lysine increases negative charge, decreasing affinity to DNA
(Remember, DNA is negatively charged)
TIghter coiling limits access for transcription factors
What does methylation of lysine do?
Methylation of Lysine decreased negative charge, increasing affinity to DNA
What does acetylation of Lysine do?
Acetylation of Lysine increases negative charge, decreasing affinity to DNA
What is Genomic Imprinting?
Genomic Imprinting: Phenomenon in which expression of an allele depends on the parent that transmits it
Methylation at imprinting control regions (ICRs) affects gene expression
RNA-Seq has identified about 100 imprinted genes
Paternally imprinted: paternal allele not transcribed
Maternally imprinted: material allele isn’t transcribed
Imprinted = silenced
If I silence a paternal or maternal allele, what happens?
RNA-Seq has identified about 100 imprinted genes
Paternally imprinted: paternal allele not transcribed
Maternally imprinted: material allele isn’t transcribed
Imprinted = silenced
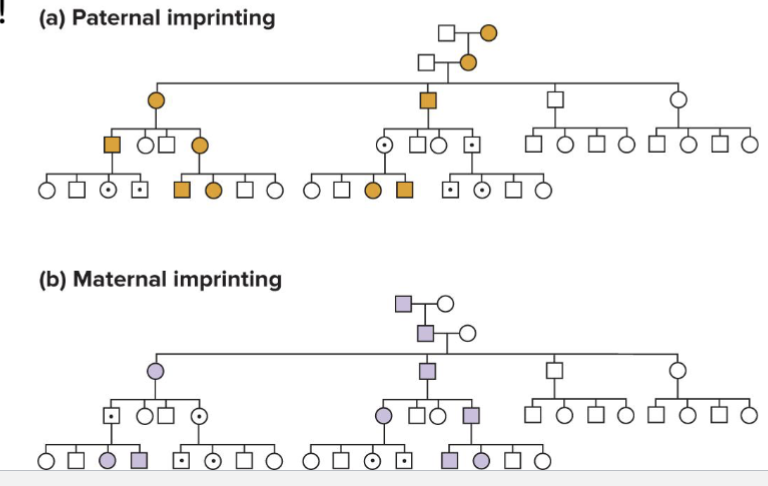
How can imprinting in pedigrees look?
Alleles from the non-imprinting parent are dominant in offspring
How was imprinting discovered?
Scientists predicted existence of imprinting prior to understanding the mechanism
In the 1980s, pedigrees were observed where the sex of the parent carrying the disease allele determined if the child had the disease
Deletion of the paternally imprinted gene doesn’t affect a father’s child if the wild type allele from the mother is expressed
Deletion of a maternally imprinted gene doesn’t affect a mother’s child if the wild type allele from the father is expressed
How can deleting paternally imprinted gene affect a child?
Deletion of the paternally imprinted gene doesn’t affect a father’s child if the wild type allele from the mother is expressed
How can deleting maternally imprinted gene affect a child?
Deletion of a maternally imprinted gene doesn’t affect a mother’s child if the wild type allele from the father is expressed
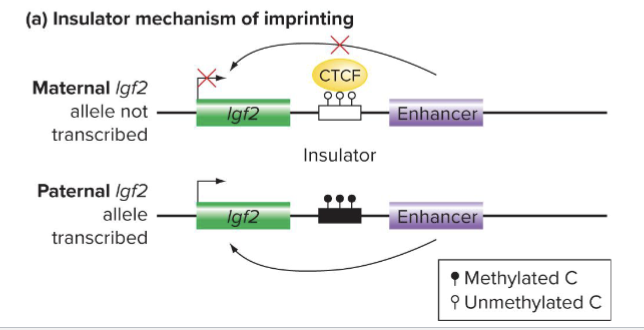
Describe Imprinting Mechanism: INSULATORS
EX: ICR is an insulator that controls transcription of Igf2 gene
Nonmethylated maternal insulator is functional,; binds CTCF and transcription is silenced
Methylated paternal insulator is nonfunctional and transcription is active
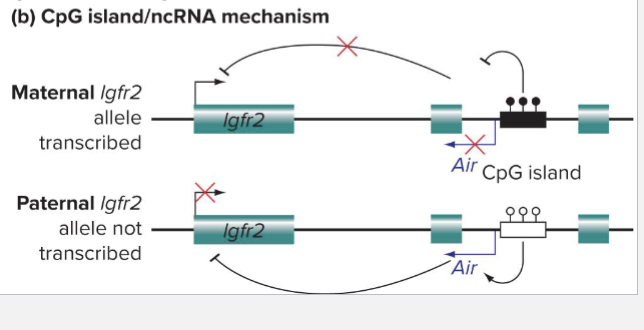
Describe Imprinting Mechanism: ncRNA MECHANISM
ICR contains a noncoding RNA whose transcription is controlled by CpG islands
Nonmethylated paternal CpG island allows production of Air ncRNA; transcription of lgfr2 is silenced
Methylated maternal CpG island prevents production of Air; transcription of lgfr2 is active
How does Resetting of Genomic Imprints work?
Epigenetic imprints remain throughout the lifespan of mammal
During meiosis, imprints are erased and new ones are set based on the sex of organism
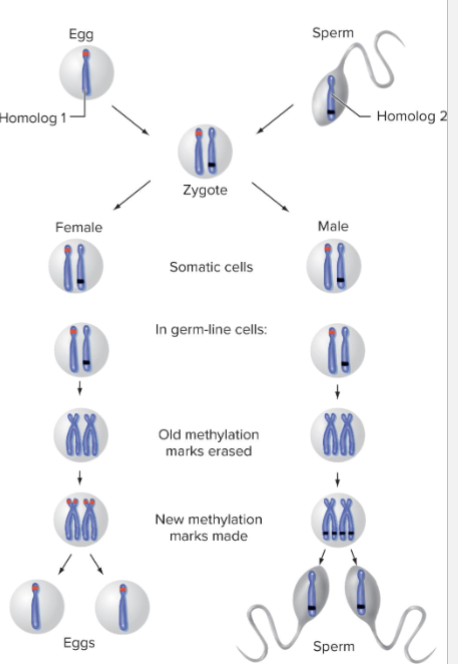
What is EXAMPLE of Resetting of Genomic Imprints?
Two syndromes associated with small deletions of Chromosome 15, q11-13 region
At least 2 genes within region are differently imprinted
Prader-Willi syndrome occurs when deletion is inherited from father
Angelman syndrome occurs when the deletion is inherited from the mother
Affected individuals have mental/development disorders
SEE IF INCORRECT DOUBLE CHECK
What are reasons for imprinting?
In placental mammals, most imprinted genes control prenatal growth
Parental conflict hypothesis: mother’s best interest for her baby to be small; father’s interest for his baby to be large and robust
Viewed as overly simplisitic, other theories considered
Most control of imprinting is maternal, so why would mother cede control to paternal allele
Alternate hypothesis is to encourage rapid adaptation by increasing selective pressure on developmental genes
What is parental conflict hypothesis?
Parental conflict hypothesis: mother’s best interest for her baby to be small; father’s interest for his baby to be large and robust
What is cell fate?
Imprinting is RARE
Many epigenetics marks are only transmitted through mitosis
All cells in multicellular organsims have the same genes
Cell types are different bc of gene expression differences
Master regulator, often a transcription factor, determines gene expression and potential fates
Cells “remember” their fate from cell generation to cell generation in part by copying histone modifications at replication fork
Nucleosomes on both daughter strands are composites of new and old histones
Example of master regulators?
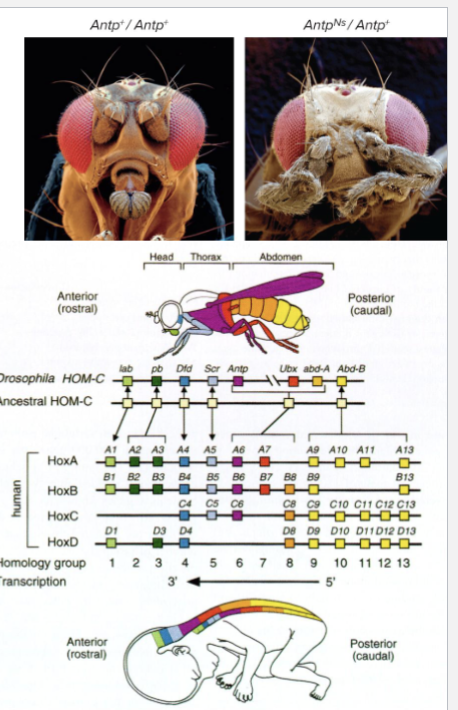
HOX GENES
Describe Hox Genes
Drosophila Hox (homeobox) genes encode transcription factors that set up animals segmented body plan
Cells in each fly segment get information about which segment they are in due to Hox proteins made there
Hox genes are repressed in specific groups of cells
HIGHLY CONSERVED
If epigenetic markers affected - likely passed down (asked in class by me)
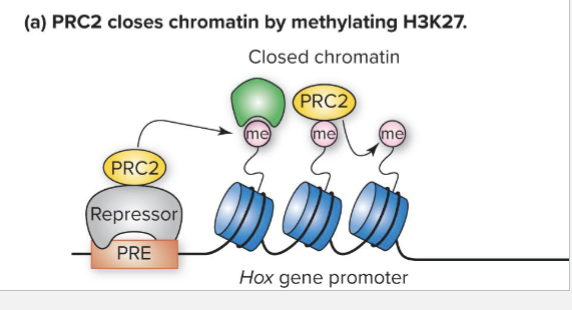
How can Hox Genes Repression work?
PRE (Polycomb response element): Cis-acting DNA sequence that binds repressors that recruit PRC2 co-repressor complex
PRC2 contains a histone methyltransferase that methylates lysine27 (K27) in the tail of histone H3
H3K27me mark is propagated to adjacent nucleosomes and Hox genes is repressed
PRC2 is recruited to H3K27me
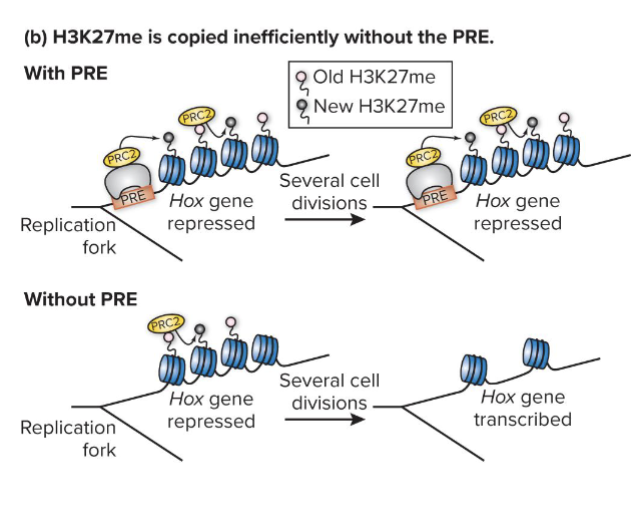
How can Hox repression be maintained?
Once established H3K27me and closed chromatin are recapitulated at the Hox gene in all of the descendants of a cell
PRC2 recruitment to PRE maintains robust silencing
H3K27me marks are self perpetuating but not indefinitely
How does Constitutive Heterochromatin work?
Constitutive Heterochromatin: highly compacted chromatin found in all cells
Found at centromeres, which are enriched in transposable elements; important to prevent TE mobilization
Centromeric heterochromatin formation often involves methylation of K9 amino acid on the tail of histone H3 (H3K9me)
How does Heterochromatin formation work in plants?
ncRNA transcribed from centromeres of plant chromosomes intiates heterochromatin formation
ncRNA is converted to siRNAs which bind to Argonaute (ago) proteins
Ago recruits histone methyltransferases (HMTs) that methylate H3K9 and DNMTs that methylate cytosine in DNA; heterochromatin forms
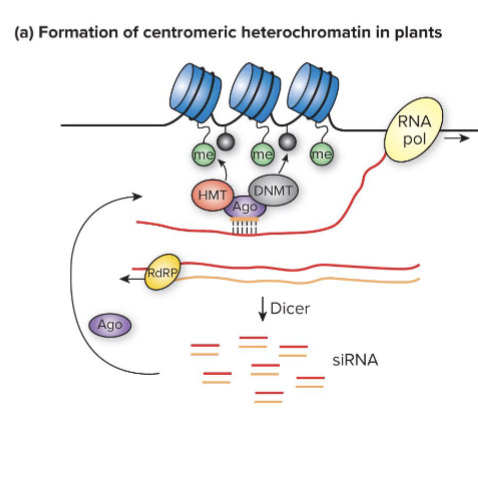
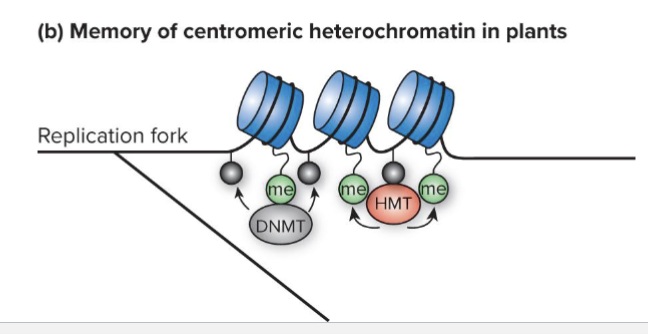
How can plant heterochromatin be maintained?
Plant heterochromatin be re-formed after cell division
H3K9me marks remaining on some nucleosomes recruit a DNMT that methylates cytosines
Methylated cytosines attract an HMT in feedback loop
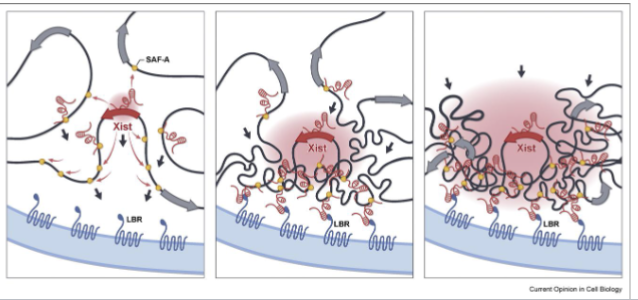
What is X-inactivation?
Cells in early female mammalian embryos inactivate a random X chromosome, making it a Barr Body
Xist IncRNA binds to X chromosome that expresses it and recruits histone modifying enzymes including PRC1 and 2
Chromatin marked by H3K9me, H3K27me, methylated CpG islands among others
How does X-inactivation spread?
Xist serves as a scaffold for various RNA binding proteins (RBPs)
SAF-A links Xist to the chromosome, while Lamin-B receptor links it to the nuclear lamina
Causes X chromosome to clump together and facilitates spread of Xist
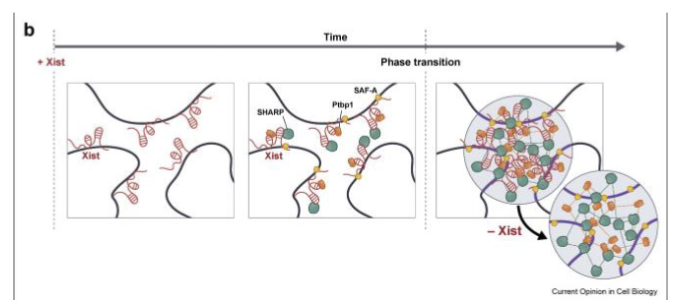
What is the heritability of X inactivation?
Not really understood
Transfer of histone marks/chromatin methylation is important but not fully explanatory
Possible that Barr body is compartmentalized enuff to maintain repressive environment during replication
What is transgenerational epigenetic inheritance?
Transgenerational epigenetic inheritance: when an environmentally induced trait (not caused by base pair mutation) appears in an individual descendants whose DNA was not directly exposed to environmental trigger
Some examples:
RNA directed DNA methylation
piRNA memories of Transposable Element (TE) invasion
Describe Toadflax Flower specification?
Wild type, bilateral, vs. Peloria, radial, flower symmetry specified by a recessive nontranscribed allele of a Lcyc gene
Same DNA sequence, but Peloria Lcyc allele is extensively methylated and silenced
Epialleles: alleles that derive from epigenetic marks instead of sequence changes
Cause is methylation of upstream TE extending into lcyc
Heritable bc DNA methylation marks aren’t erased in plant gametogenesis

What is piRNAs?
Organisms try to limit TE mobilization to prevent mutations/chromosomal rearrangements
In Drosophila, small non-coding RNAs called piRNAs (Piwi-interacting RNAs) suppress TE movement
piRNAs are made in female germ-line cells and transmitted to progeny in oocytes
Mechanism to preserve memories of TE invasions and help protect against future TE mobilization
What is piRNA mechanism?
TE invading Drosophila genome hops into piRNA cluster, which contains inactive remnants of different TEs
Cluster is transcribed to make piRNA precursors
piRNAs promote destruction of TE mRNAs blocking expression of TE genes and immobilizing TEs
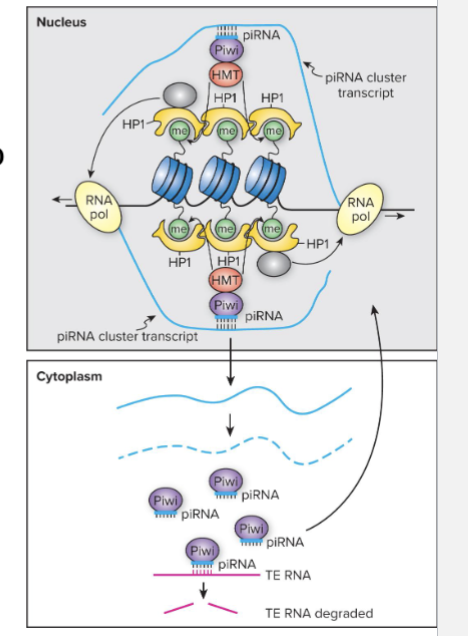
How does piRNA inheritance work?
piRNAs can regenerate themselves in the female germ line
piRNAs guide the Argonaute protein piwi back to the same piRNA cluster by basepairing with piRNA transcripts as they are being transcribed
Piwi recruits histone modifying enzymes
HP1 binds to histone and closes chromatin, but the HP1 at piRNA clusters brings RNA polII to clusters, leading to transcription
Self-sustaining germline production of piRNAs
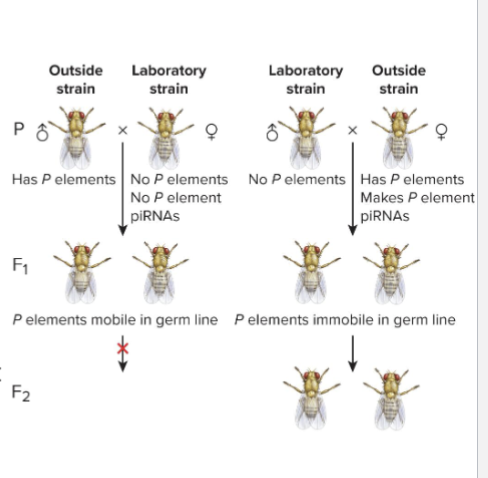
What is Hybrid Dysgenesis?
STERILITY of hybrid progeny
Outside flies crossed to lab Drosophila had STERILE progeny
But only when outside males were crossed with lab females and not reverse
Genomes of outside flies contained P elements; lab fly genomes had none
Oocytes of females w/out P element don’t have piRNAs to press P element mobilization
Leads to so many mutations in gametes that progeny are sterile
Oocytes of females with P elements have piRNAs that inactivate P elements in germ line of progeny
What is Genomics?
Human Genome Project: an accurate sequence of the human genome was initiated in 1990 and completed (97% converage) in 2003
We’ve now seqeucned thousands of eukaryotic and hundreds of thousands of prokaryotic species
General ideas behind genome sequencing are simple:
Fragmenting the genome
Cloning DNA fragments
Sequencing DNA fragments
Reconstructing genome from fragments
What are Restriction Enzymes?
Restriction Enzymes fragment the genome at specific sites by recognizing a specific sequence of bases
Cuts sugar-phosphate backbones of both strands
Restriction fragments are generated by digestion of DNA with restriction enzymes
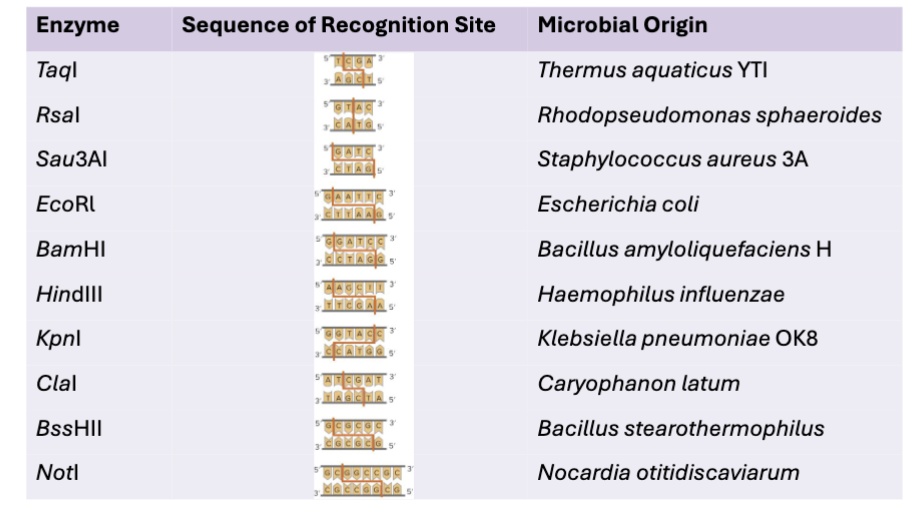
Hundreds of restriction enzymes now available
Recognition sites for restriction enzymes are usually 4 to 8 bp of double strand DNA
Often palindromic - base sequences of each strand are identical when read 5’ to 3’
Each enzymes cuts at the same place relative to its specific recognition sequence
What are some commonly used restriction enzymes?
What are Blunt vs. Sticky ends?
Blunt ends: cuts are straight through both DNA strands at the line of symmetry
Sticky ends: cuts are displaced equally on either side of line of symmetry
Ends have either 5’ overhangs or 3’ overhands
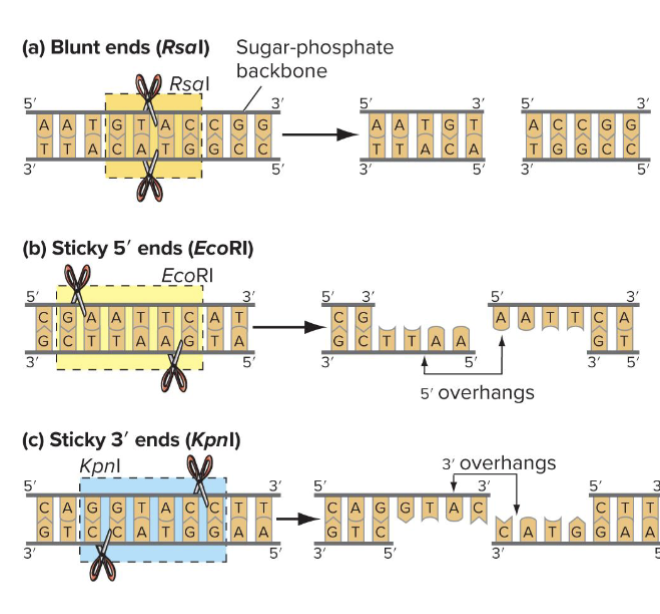
Different restriction enzymes produce different fragment length - calculate?

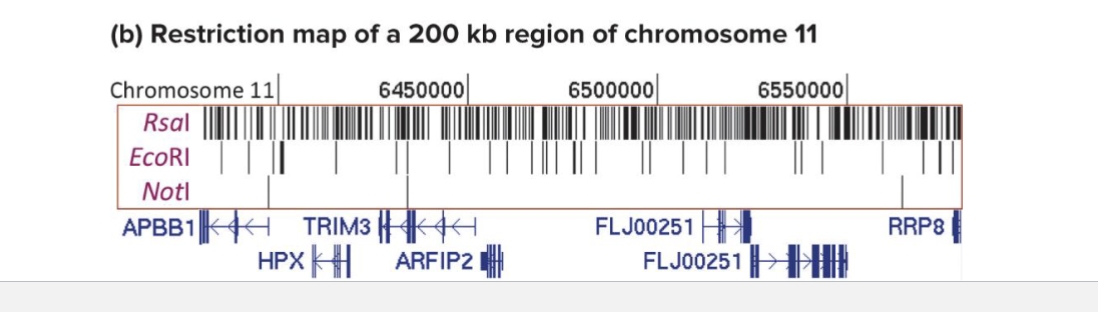
What is an example of a restirciton map?
Site of 3 restirction enzymes in 200 kb region of human chromosome 11
Names and locations of genes in this region are shown below restriction sites
Rsal 4bp, EcoRI 6bp, Notl: 8bp
What is Mechanical Fragmentation?
Some experiments require random cutting of DNA, not guided cuts with restriction enzymes
Mechanical forces can break phosphodiester bonds
Passing DNA through thin needle at high pressure
Sonication (ultrasound energy)
Ends can be blunt, may have protruding single-strand regions
How to do Gel Electrophoresis?
DNA analysis via electrophoresis, movement of charged molecules (ex: DNA fragments) in electric field
Steps:
1) Pour heated molten agarose into acrylic plate to which comb has been attached allow to cool and harden
2) Remove comb, place gel in buffered aqueous sol, load DNA samples into wells in gel
3) Apply electric current
4) DNA has negative charge, so moves toward postitive charge
5) Remove gel from tank after electrophoresis
6) Visualize DNA fragments by staining gel with fluorescent dye, photograph gel under UV light
With linear DNA fragments migration distance through gel depends on size
Determine size of unkwown fragments by comparison of migration to DNA markers of known size
How can DNA fragments be cloned?
Genomes of animals, plants, and microorganisms are too large to analyze all at once
Molecular cloning purifies a specific DNA fragment away from all other fragments and makes identical copies of fragment
2 basic steps:
Insert DNA fragments into cloning vectors to specialized chromosome-like carriers that ensure transport, replication, and purification of DNA inserts
Transport recombination DNA into living cells to be copied
Group of replicated DNA molecules = DNA clone
What as Plasmid Vectors?
Plasmid cloning vectors have 3 main features:
Origin of replication
A selectable marker gene (ex: antibiotic resistance)
Insertion site
Often a synthetic polylinker, DNA sequence containing multiple restriction enzyme sites
Different vectors carry different size insertions
Plasmid ~20 kb
Bacterial artificial chromsomes (BAC) ~~ 300 kb
Yeast artificial chromosomes (YAC) ~~ 2000 kb
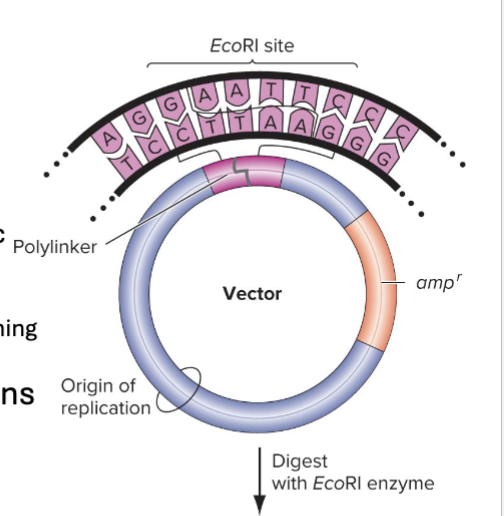
How can recombinant vectors be created?
Digest the vector and target genomic DNA with a restriction enzyme that results in complementary sticky ends
DNA Ligase is used to seal the phosphodiester backbones between vectors and insertion
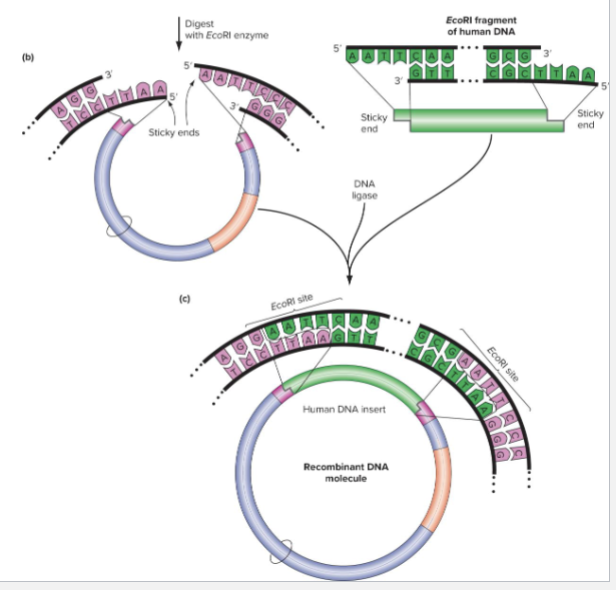
Each genomic DNA fragment can form _____?
DIFFERENT RECOMBINANT MOLECULE
Pieces being inserted are random
Result is many different recombinant vectors
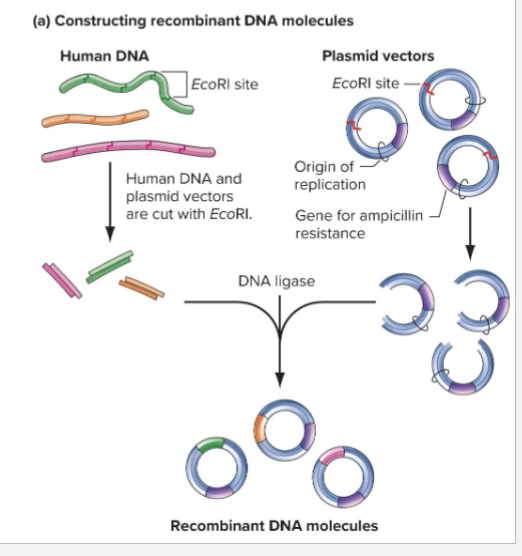
What can you do with recombinant DNA to transform Host cells?
Transformation: process by which cell or organism takes up foreign DNA
In E.coli only 0.1% of cells will be transformed with plasmid
Only cells with plasmid will grow on media with ampicillin
Each cell with plasmid will produce a colony on agar plate, millions of identical plasmids in colony are a DNA clone
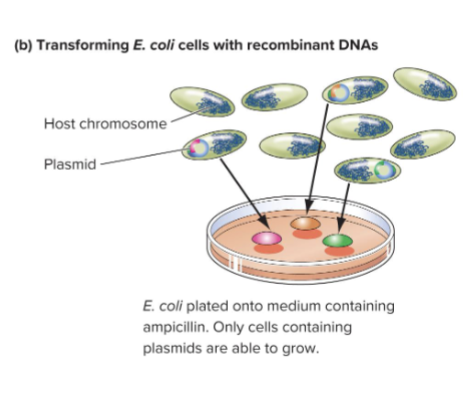
What is a Genomic Library?
Genomic library: long lived collection of cellular clones that contains copies of every sequence in the whole genome inserted into a suitable vector
Each colony contains a different recombinant plasmid, each with a part of the genome
A perfect genomic library has one copy of every sequence in the entire genome
Genomic Equivalent: Minimum # of clones in a perfect library = length of genome/length of insert
Impossible to obtain perfect library
Usually libraries are made that have 4-5 genomic equivalents
Gives an average of 4-5 clones for each locus (95% prob that each locus is present at least once)
What is Sanger Sequencing?
Uses DNA polymerase to synthesize new DNA
DNA polymerase requires:
TEMPLATE: single strand of DNA to copy
Deoxyribonucleotide triphosphates (dATP, dCTP, dGTP, dTTP), basic building blocks for new DNA
Primer: short single stranded DNA molecule that is complementary to part of template, includes free 3’ end to which DNA polymerase can attach new nucleotides
What are Sanger Sequencing Steps?
Cloned recombinant DNA is denatured (heat breaks H bonds btw strands)
Strands are mixed with oligonucleotide primer (made in DNA synthesizer) approximately 20 bp complementary to template strand
As temperature is lowered, primers and template strand anneal (hybridize)
Hybridized template and primers are mixed with DNA polymerase, dNTPs, and small amounts of dideoxyribonucleotide triphosphates, ddNTPs
ddNTPs lack a 3’-OH, halting polymerization
Modtern method has each with unique florescent tag
Result is a set of nested fragments, with different 3’ ends
How can you readout the Sanger Sequencing result?
Special gel seperates DNA fragments that can differ in size by only one nucleotide
Smaller DNA fragments migrate faster
Each lane displays the sequence obtained form a separate DNA sample and primer
Each fragment terminates with a specific ddnTP labeled with unique florescent dye
DNA fragments are electrophoresed based on size, color of terminal ddNTP is recorded to identify nucleotide code
How can sequencing reads be automated?
Fragments flow past a laser beam and the color of the terminal base is digitally recorded
Computer reads of sequence complementary to the template strand
Sequence is read from left to right (5’ to 3’ synthesis from primer)
Genome comparisons?
How can genomes be assembled (Describe Human Genome Project Method)
Human Genome Project
1) Construct BAC genomic library
2) Identify sets of overlapping BAC clones
3) Shear DNA from each BAC seperarely to make smaller clones
4) Sequence DNA
5) Assemble sequences based on overlap
Strategy was successful but labor intensive, inefficient and expensive
How does Shotgun sequencing work?
Celera developed the whole genome shotgun strategy for genome sequencing
Create genomic library of overlapping fragments in plasmid vectors
Sequence DNA inserts of randomly chosen library plasmids (“shotgun”)
Assembly sequences based on overlap of sequences into contigs - continuous base pair sequences
The whole-genome shotgun approach can be highly automated
Can repeats disrupt or help shotguns sequencing?
DISRUPT
MAny sequences repeat throughout genome
With only short, single reads these sequences cannot be distinguished from each other
Can lead to misassembly
How does Paired End Reads work?
Paired end sequencing, sequencing a BAC clone library rather than smaller inserts of plasma clones
Each BAC clone insert gave 3 pieces of into:
2 approximately 1000 bp sequence reads
Knowledge that 2 sequences were approx 200-300 kb apart
Can give information on flanking sequences of repeat
What is Next Generation Sequencing?
Sequencing an entire genome (high coverage, missing some small regions) now costs about $1000
Whole-exome sequencing (limited to expressed parts of genome) is less expensive
Sequence only fragments corresponding to exons
High-throughout, or massively parallel, sequencing is like Sanger sequencing with a few modifications
Individual DNA molecules are anchored in place
Each base is identified before next one added
Increased sensitivity eliminates need for cloning or PCR
What is ILLUMINA SEQUENCING: LIGATION
Cloning into vectors is time consuming so Illumina sequencing skips it
Instead of ligating into vectors, ligates with adaptors that have
Sequencing and amplification primer sites
Complementation to DNA molecules anchored to flow cells
Sometimes a barcode (unique seq. identifier) to ID source of sequence
What is ILLUMINA SEQUENCING: AMPLIFICATION
Anchorign primers used to synthesize new strands
After denaturing the other end is bound by reverse primers also anchored to flow cell
Synthesizes new strand
Repeat process
Cleave 1 strand type
Result is clusters of identical sequences
What is ILLUMINA SEQUENCING: SEQUENCING
Add sequencing primers
Add Fluorescently tagged bases with blocked 3’OH and DNA polymerase
Read Fluorescent label at each cluster
Remove fluorophore and blocker
Repeat
Reads 50-200bp
What is Illumina Use Cases?
Illumina sequencing is best used if reference sequence available
Not ideal for de novo whole genome sequencing
This way the 50-200 bp fragments can be aligned to known sequences
Good for sequencing whole genomes of humans