Structural Biology - Alpha Helix and Domain Structure
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
50 Terms
Short Answer: It is generally observed that polar and hydrophilic main chain atoms tend to remain on the protein surface. Yet, they are sometimes found buried in the protein interior. How is this possible?
Polar and hydrophilic main chain atoms can be buried inside the protein if they are involved in internal hydrogen bonding, such as within α-helices, β-sheets, or other secondary structures. These hydrogen bonds stabilize the polar atoms and compensate for the loss of interactions with water.
Short Answer: What secondary structure is associated with phi ≈ -57° and psi ≈ -47°?
Right-handed α-helix. These angles fall in the lower left region of the Ramachandran plot.
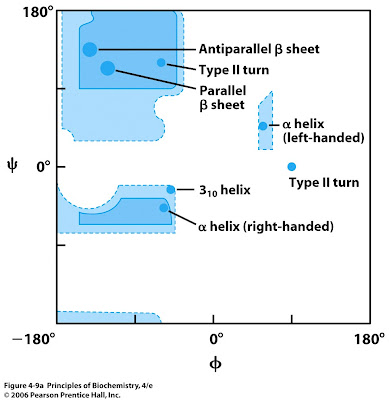
Short Answer: Explain what is a Ramachandran plot. What does it help with when it comes to novel protein structures?
A plot that shows the allowed phi and psi angles to construct a secondary structure. This comes especially handy when verifying novel protein structures.
Short Answer: What secondary structure corresponds to phi ≈ -139° and psi ≈ 135°?
β-sheet (antiparallel). These angles are located in the top left quadrant of the plot.
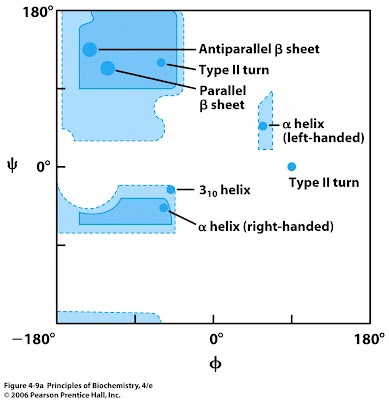
Short Answer: Which amino acid is the most restricted in its allowed phi and psi angles?
Proline. Its cyclic structure limits backbone flexibility.
Short Answer: Which amino acid is the most flexible in terms of phi and psi angles?
Glycine. It lacks a side chain (only H), allowing it to occupy more of the Ramachandran space.
Short Answer: Why are left-handed α-helices not commonly found in proteins, while right-handed ones are more favored?
Left-handed α-helices are less favored due to unfavorable steric interactions.
Short Answer: Which quadrant of the Ramachandran plot corresponds to the right-handed α-helix? Their values?
Bottom left. Typical values: φ ≈ -57°, ψ ≈ -47°.
Short Answer: Which quadrant of the Ramachandran plot corresponds to antiparallel β-strands? Their values?
Top left. Typical values: φ ≈ -139°, ψ ≈ 135°.
Short Answer: Which quadrant of the Ramachandran plot corresponds to parallel β-strands? Their values?
Top left. Typical values: φ ≈ -119°, ψ ≈ 113°.
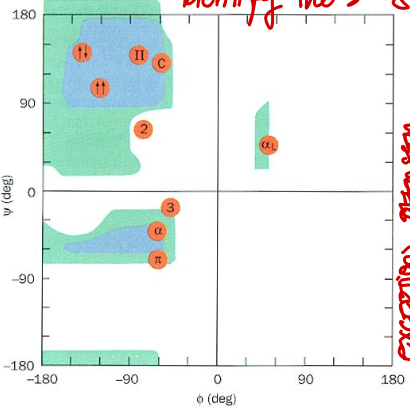
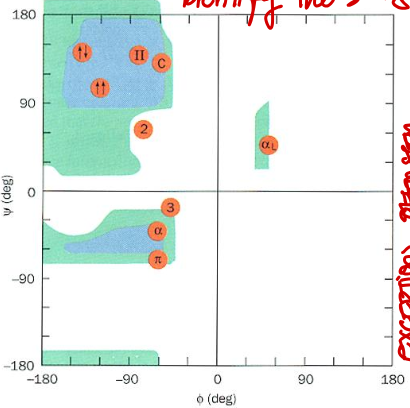
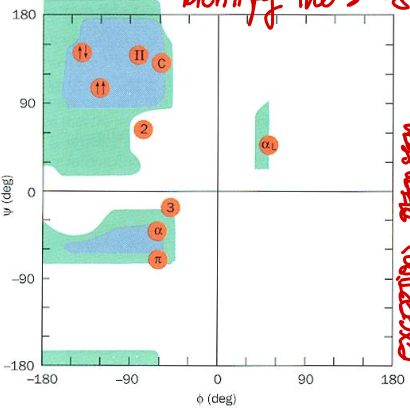
Short Answer: Which quadrant of the Ramachandran plot corresponds to right-handed 310 helix? Their values?
Top left. Typical values: φ ≈ -49°, ψ ≈ -26°.
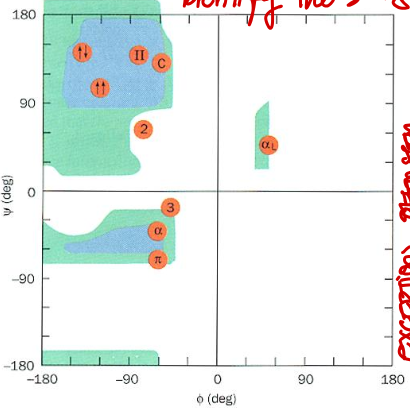
Short Answer: Which quadrant of the Ramachandran plot corresponds to right-handed π helix? Their values?
Bottom left. Typical values: φ ≈ -57°, ψ ≈ -70°.
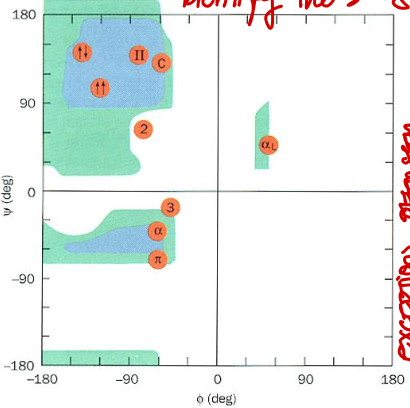
Short Answer: Which quadrant of the Ramachandran plot corresponds to 2.27 ribbon? Their values?
Top left. Typical values: φ ≈ -78°, ψ ≈ 59°.
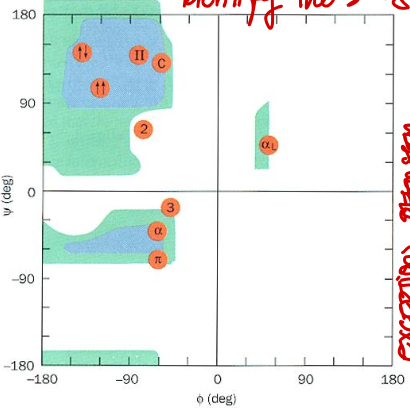
Short Answer: Why are certain regions of the Ramachandran plot disallowed?
Answer: Due to steric clashes between atoms, which make some phi/psi combinations energetically unfavorable.
True or False: D-amino acids would produce a mirrored Ramachandran plot compared to L-amino acids.
True. The plot is mirrored across the origin.
Short Answer: You observe several residues in a newly solved crystal structure with phi/psi angles in disallowed regions of the Ramachandran plot. What might this indicate about the model?
It could suggest errors in the structural model, such as poor refinement, misfit backbone, or incorrect side-chain rotamers affecting backbone geometry. Ramachandran outliers are commonly flagged in structural validation reports.
Short Answer: A glycine residue is found in a region of the Ramachandran plot normally disallowed for other residues. Is this acceptable? Why or why not?
Yes. Glycine lacks a bulky side chain, allowing more backbone flexibility and access to otherwise disallowed regions.
Short Answer: In a mutation study, a glycine in a tight turn is replaced with valine. How might this affect the local backbone conformation?
Answer: The valine introduces steric bulk and limits phi/psi flexibility, potentially disrupting the turn or causing strain due to inability to adopt glycine-like angles.
Short Answer: In a misfolded protein model, most residues lie in allowed Ramachandran regions, but the protein is non-functional. What does this suggest about the Ramachandran plot’s limitations?
The plot checks local backbone geometry, but doesn’t account for global folding or functional conformation—so a model can be geometrically valid but biologically incorrect.
Short Answer: In naturally occurring proteins, α-helices are always right-handed. Why are left-handed helices not observed in real protein structures, despite being theoretically possible?
Answer: Right-handed helices are favored due to the chirality of L-amino acids, which makes the right-handed conformation sterically and energetically more favorable.
Short Answer: Which of the α-, 3₁₀-, and π-helices is the most common in proteins?
α-helix. It is the most stable and most frequently occurring helix in protein secondary structure.
True or False: The 3₁₀-helix is more tightly wound (narrower) than the α-helix.
True. It has fewer residues per turn and a smaller radius.
Short Answer: What is the hydrogen bonding pattern in an α-helix?
i → i+4. The backbone C=O of residue i hydrogen bonds with the N-H of residue i+4.
Short Answer: What is the hydrogen bonding pattern in a 3₁₀-helix?
i → i+3. This tighter bonding pattern leads to a narrower helix.
Short Answer: What is the hydrogen bonding pattern in a π-helix?
i → i+5. This creates a looser, wider helix.
Short Answer: Rank the helices from most to least tightly wound.
3₁₀-helix > α-helix > π-helix (tightest to loosest).
Short Answer: What is the structural reason why 3₁₀-helices and π-helices are generally less stable than α-helices?
3₁₀-helices are less stable because their tight turn (i → i+3 hydrogen bonding) leads to steric crowding and torsional strain. Although their hydrogen bonds are more linear (and theoretically stronger), the backbone strain makes them less favorable. π-helices are less stable because their loose turn (i → i+5 hydrogen bonding) results in weaker hydrogen bonding and poorer side-chain packing, leading to cavities and instability in the core. α-helix strikes a balance between optimal hydrogen bonding, torsional angles, and efficient side-chain packing.
Short Answer: π-helices are rare compared to α-helices. When they are present, where in a protein are they typically found, and what is their functional significance?
π-helices are often found near functional sites of proteins. Their unusual geometry introduces kinks, cavities, or flexible distortions in α-helices, which can create space for substrate binding or alter local environment for catalysis.
Short Answer: What is a helical transition in protein secondary structure?
A helical transition refers to a local change in helix type along the backbone of a protein — for example, a shift from α-helix to 3₁₀-helix or from α-helix to π-helix — due to variations in backbone dihedral angles or insertions/deletions.
Short Answer: What is the function of helical transition in protein secondary structures?
Functionally, these transitions can accommodate mutations or trigger conformational changes. For example, a mutant hemoglobin protein may contain an insertion mutation. A alpha → 310 transition helps neutralize the single amino-acid insertion.
Short Answer: What types of mutations are most commonly associated with helical transitions?
Insertion or deletion of a single residue within an α-helix, causing a shift that can locally convert α-helical geometry into π-helix (insertion) or 3₁₀-helix (deletion).
True or False: Helical transitions are common across all proteins.
False. They are relatively rare and usually localized, but when present, they tend to be functionally important.
Short Answer: Why are π-helices often associated with active or functional sites despite being unstable?
Their wider diameter and kinked geometry can introduce structural flexibility, accommodate larger side chains, or enable specific binding interactions in functional regions.
Short Answer: Why are proline and glycine generally excluded from α-helices?
Because proline and glycine both act as helix breakers. Glycine has a small side chain that supports other conformations, while proline is too rigid that it introduces a kink.
Short Answer: Why do α-helices tend to favor amino acids with long and thin side chains that don’t form hydrogen bonds, like alanine and argenine?
Since these amino acids are long and slender, minimizing steric clashes with other residues. Additionally, side chain that don’t form hydrogen bonds avoid interfering with the helix’s regular i → i+4 backbone hydrogen bonding pattern, preserving stability.
Short Answer: Which amino acid has the highest α-helix-forming propensity?
Alanine. Its small, unbranched side chain (methyl group) offers minimal steric hindrance and doesn’t compete for hydrogen bonding.
Short Answer: Which amino acids are considered α-helix breakers? Name at least two.
Answer: Proline (rigid, lacks N-H for H-bonding) and glycine (too flexible, entropically unfavorable). Both disrupt the helix’s regular H-bonding pattern.
Short Answer: Which amino acids are most favorable in β-sheets?
Valine, isoleucine, and phenylalanine. These are bulky or β-branched, which are better accommodated in the extended β-sheet conformation.
True or False: Glycine is favorable in β-sheets due to its flexibility.
False. While its flexibility allows it in turns and loops, it is usually destabilizing in structured regions like helices or sheets.
Short Answer: Why do β-branched amino acids like valine and isoleucine favor β-sheets over α-helices?
Their side chains are bulky near the backbone (β-carbon), which causes steric clashes in α-helices but fit well in the extended β-sheet geometry.
Short Answer: Name one polar amino acid that is disfavored in α-helices and explain why.
Asparagine — its side chain can form competing H-bonds with backbone carbonyls, disrupting the helix’s internal hydrogen bonding.
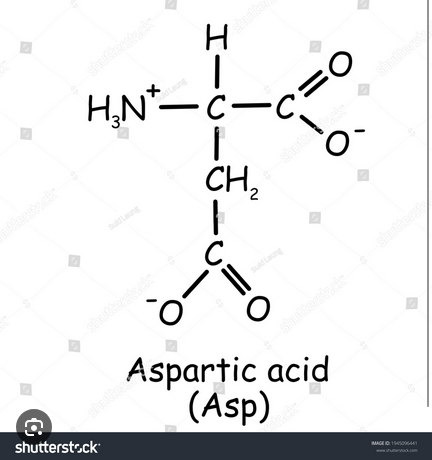
Short Answer: Name the 3 factors that lead to a bend in the alpha-helix.
Proline residues. Duh.
Packing of helices against other secondary structures in the protein core.
Exposed helices are usually bent away from the solvent region, because exposed CO groups tend to point towards solvent to maximise their H-bonding capacity.
Short Answer: What is an amphiphilic (or amphipathic) α-helix?
It is an α-helix in which one face is lined with hydrophobic residues and the opposite face with hydrophilic or charged residues. This duality allows it to interact with both hydrophobic and aqueous environments.
True or False: Glycine is often used as a helix cap at the C-terminal end due to its flexibility.
True. Its conformational freedom allows unusual torsion angles needed for effective capping.
Short Answer: Which amino acids are most commonly found at the N-cap of an α-helix?
Asparagine (Asn), Aspartate (Asp), Serine (Ser), and Threonine (Thr). These residues have side chains capable of forming stabilizing hydrogen bonds with backbone atoms at the N-terminal end of the helix.
How many residues would typically span the non-polar interior of the phospholipid bilayer?
20 residues
Short Answer: What are the two main principles that dominate the way α-helices associate in proteins?
Maximization of van der Waals packing among hydrophobic side chains: Helices associate to optimize close packing between nonpolar residues, minimizing voids and maximizing stabilizing interactions.
Helices retain conformations close to their minimum free energy when isolated: Packing does not drastically distort the helix; instead, helices interact in ways that preserve their low-energy backbone conformations.