L1: Architecture of the nucleus 1
1/63
Earn XP
Description and Tags
intro, DNA nucleosomes and chromatin
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
64 Terms
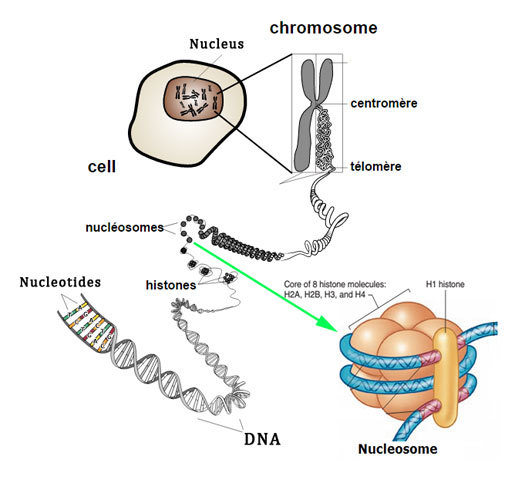
How much DNA contained in nucleus
2 metres
What is the inside the nucleus
chromatin
equal amounts of protein and DNA
→ to be able to see it
attracts dye easily which is fab
this is why chromatin is called chromatin 'chrome=colour
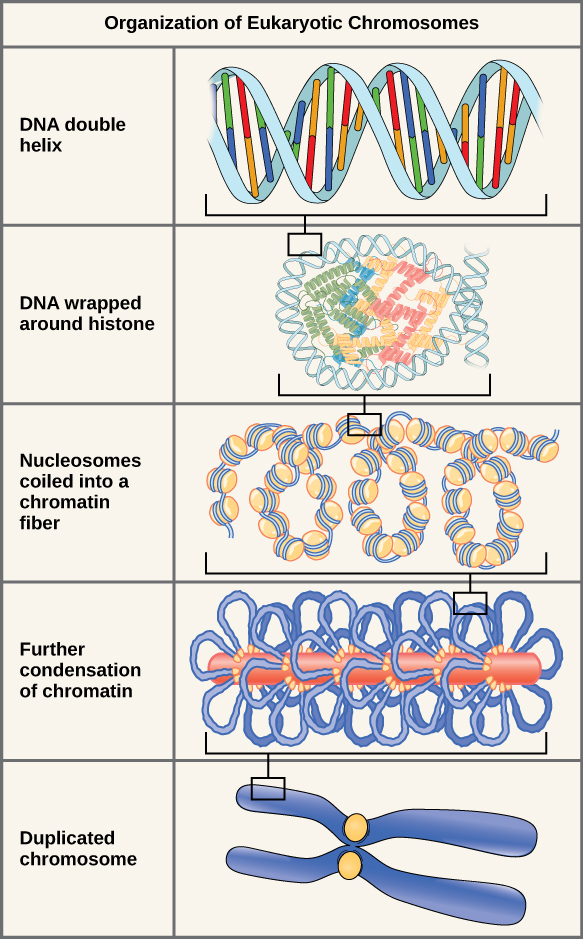
How small is the nucelus (where DNA contained)
10 um
great degree of compaction
hierachy of structure
How is it so compact?
genomic DNA associates with strucutrual chromatin components
histone proteins
forms ordered strucutres: nucleosomes
Further compaction of nucleosomes
form higher order strcutures
form chromosomes
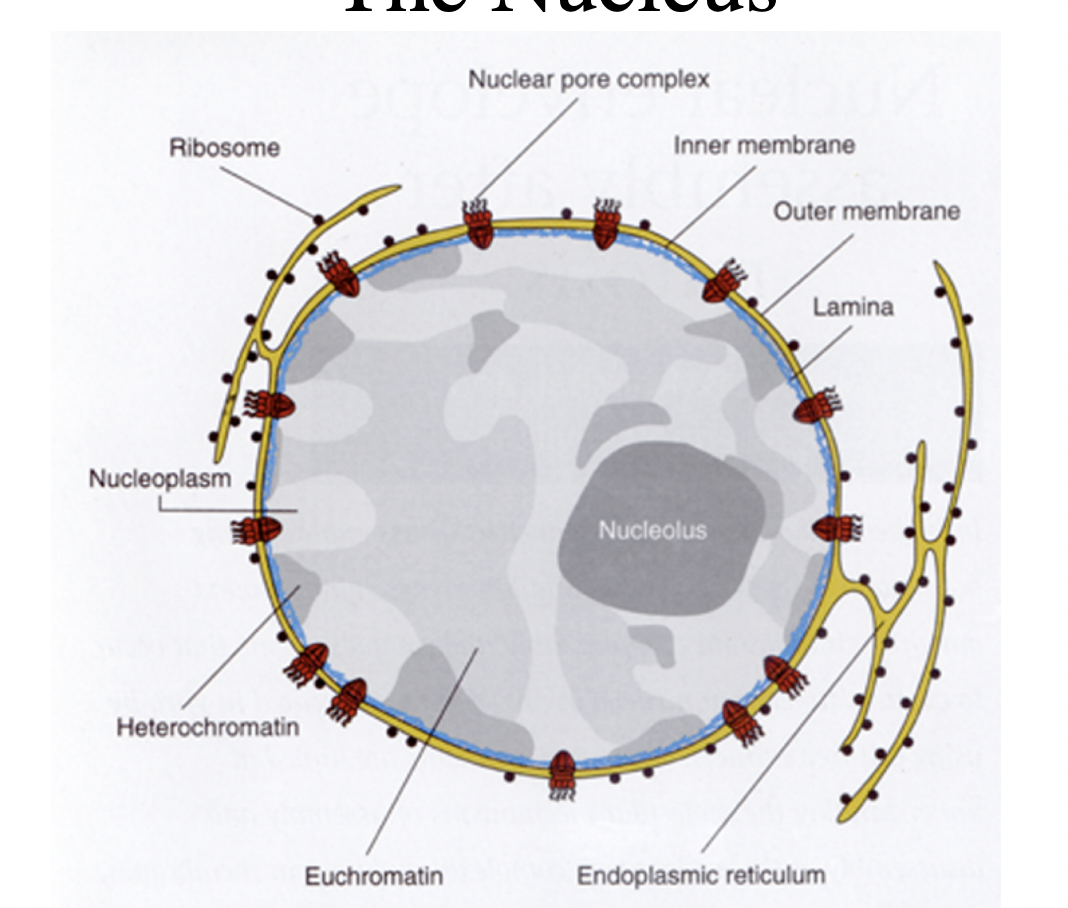
What is around chromatin?
surrounded by double lipid bilayer membrane system
forms nuclear membrane
supported by protein meshwork: nuclear envelope
What is the role of nuclear pores
regulated transport of macromolecules in and out
Role of the nucleus
information centre of the eukaryotic cell
synthesise mRNA for protteins
→ must be exported into cytoplasm for directing protein synthesis
replicate its entire strucutre accurately during each cell division cycle
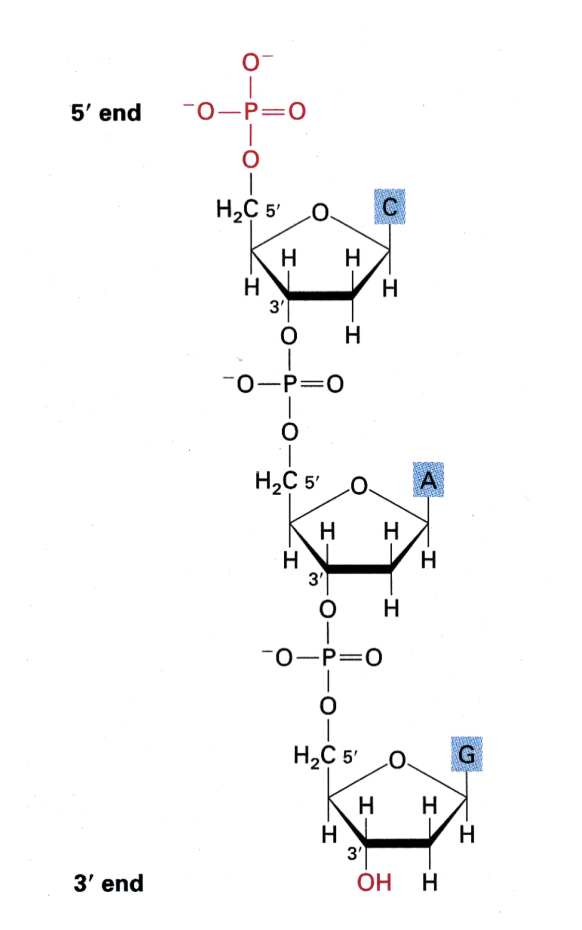
Nucleotide building blocks
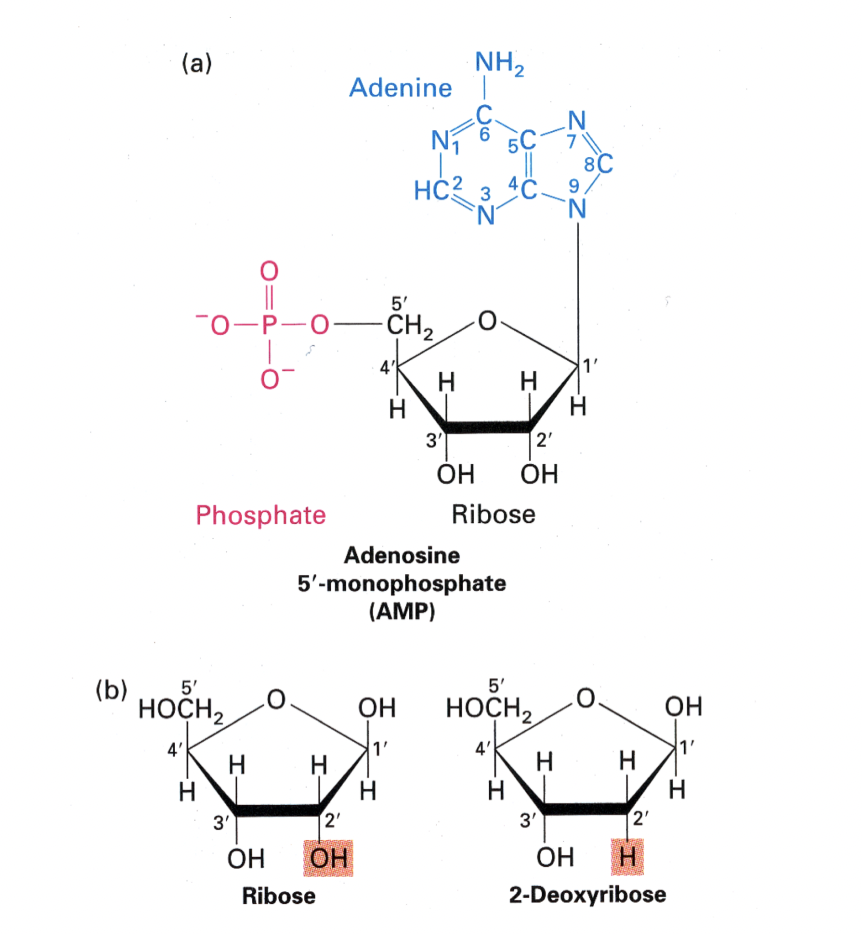
Forming the phosphodiester backbone
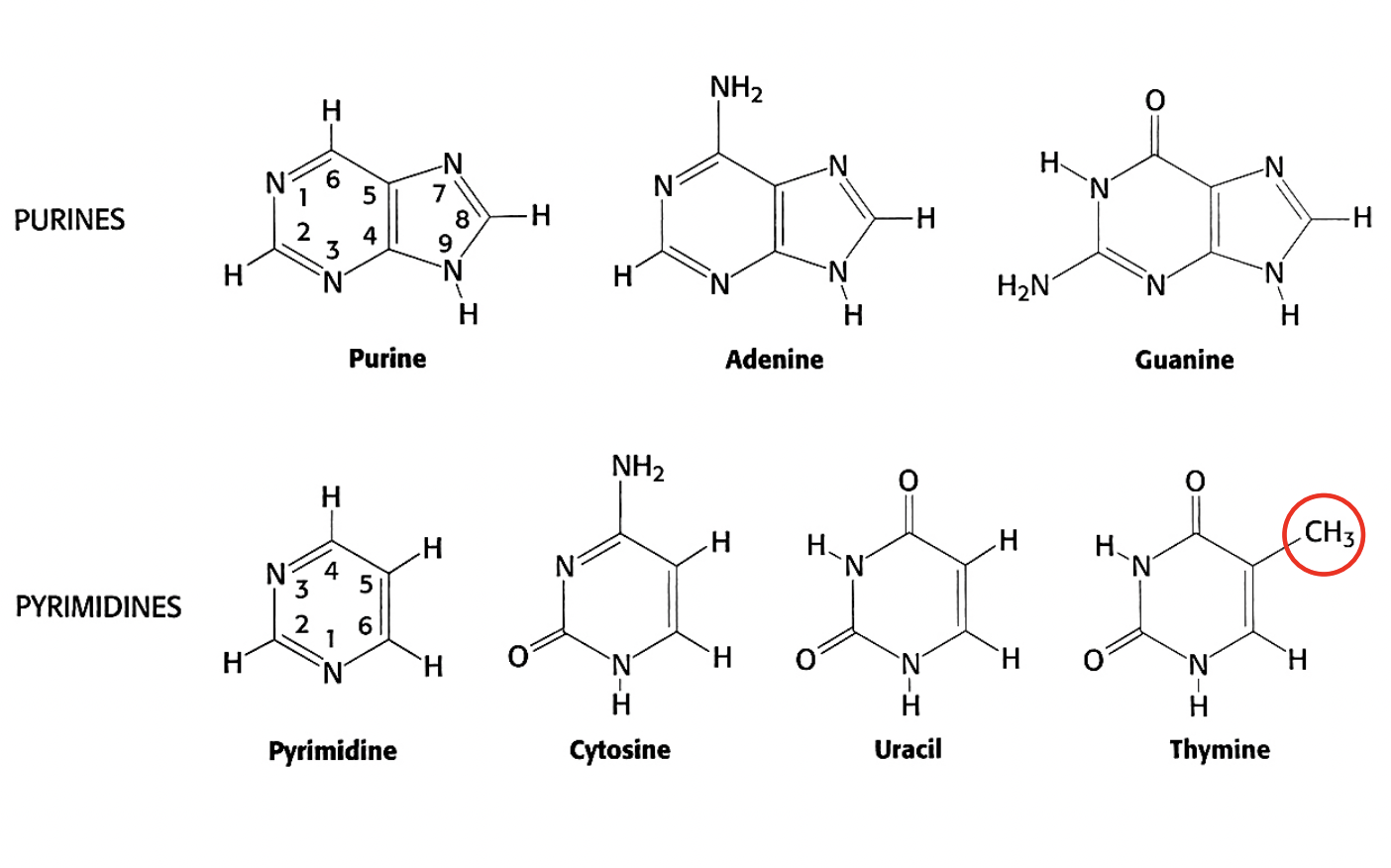
The different bases
either derived from Purine or Pyrimidine
with some modifications
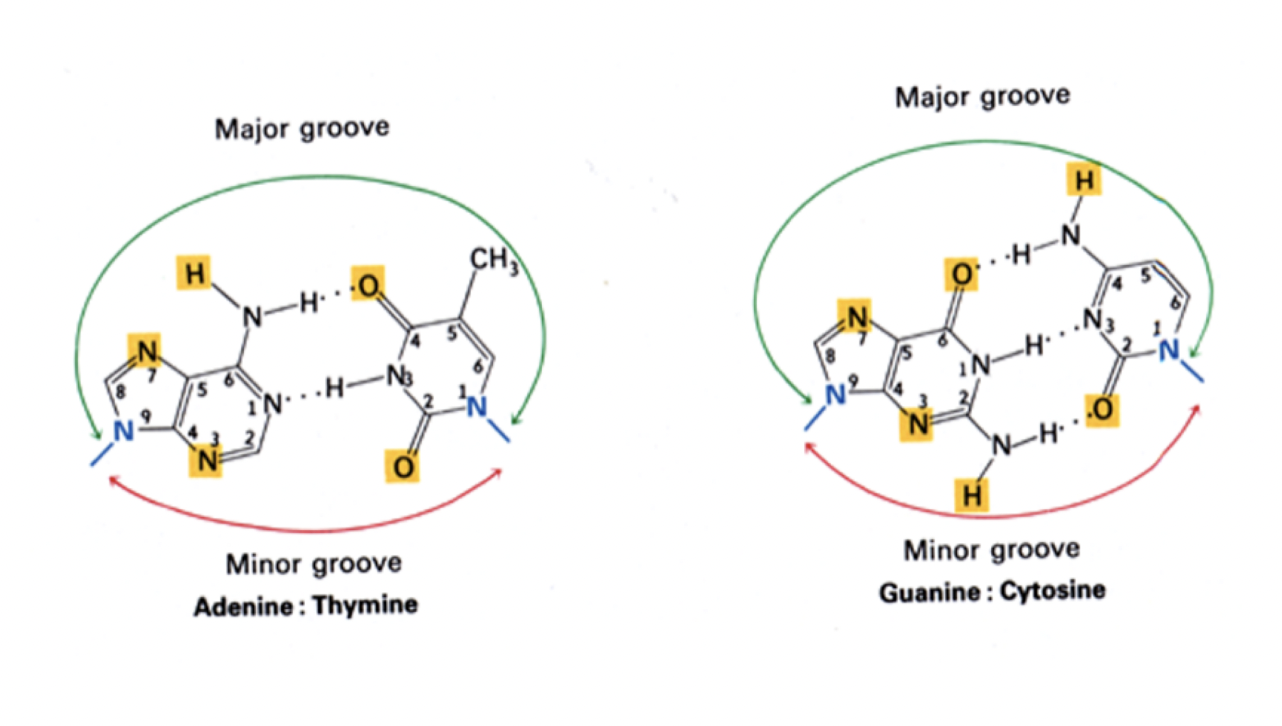
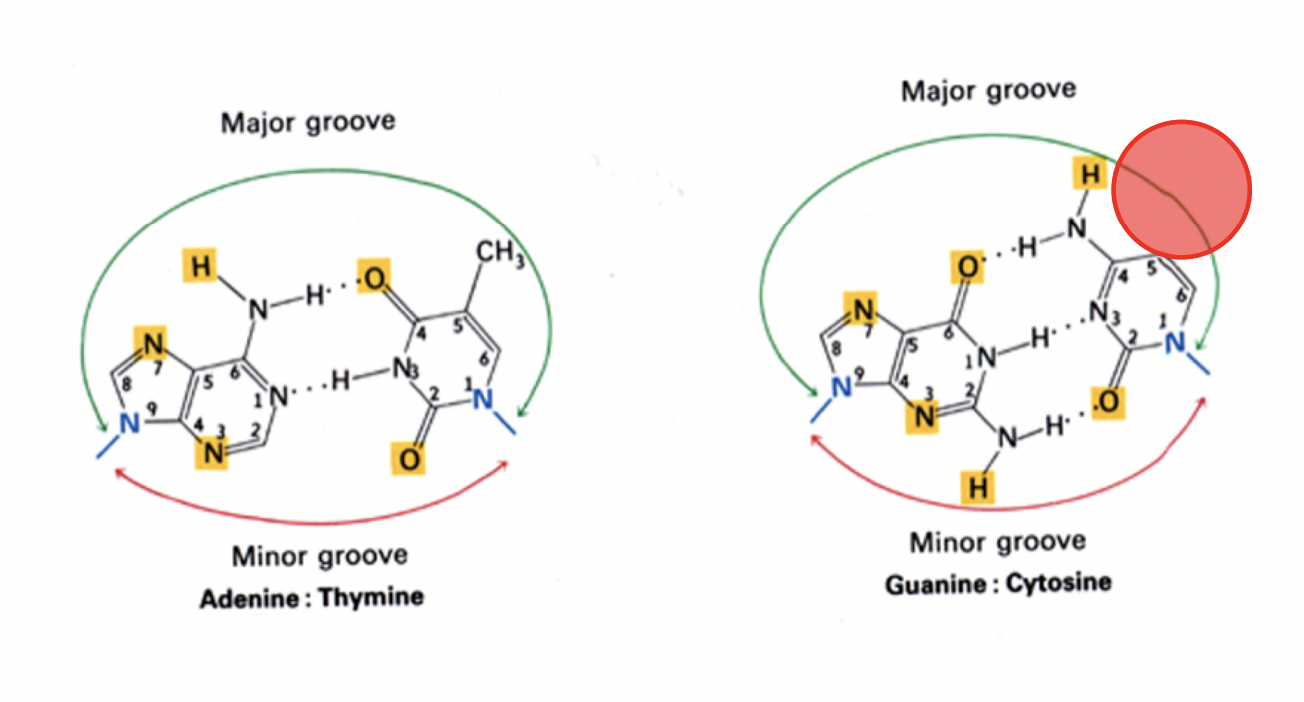
How these Base pairings make the grooves of the helix
the major and minor grooves are important in gene expression and epigenetics
DNA and its strucutre: B form DNA wehn forms
Right-handed helix
When?
physiologyical pH and salt concentration
What is the B-form of DNA? (skeletal model)
outer sugar-phosphate backbone chains connected via hydrogen-bonded base-pairs
forms major and minor groove
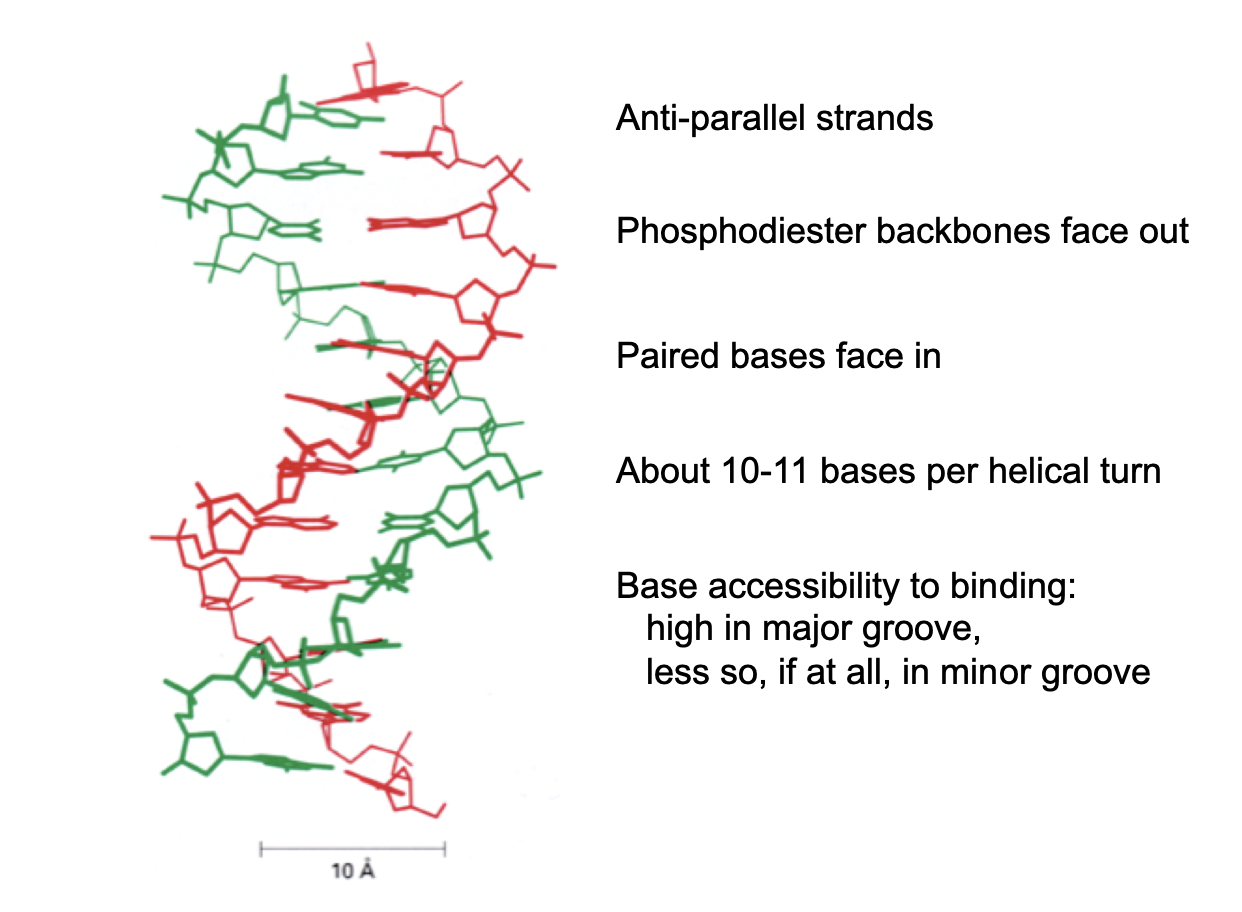
anti-parallel strands
phosphodiester backbones face OUT
paired bases face in
10-11 bases per helical turn
Base accessibility in binding
high in major groove
less so if at all in minor groove
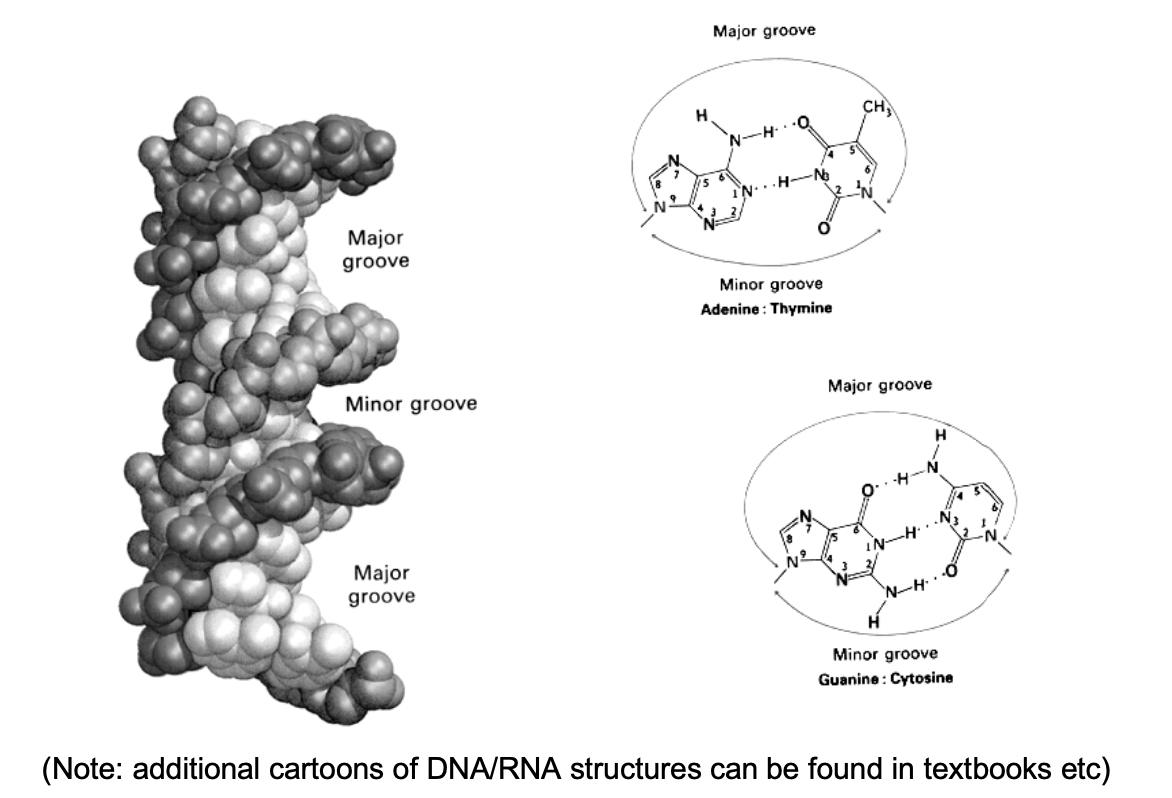
(Role of grooves)Access to genetic info is via
base-pair specific surface of the DNA helix accessible in grooves
needed for sequesnce -specific binding proteins
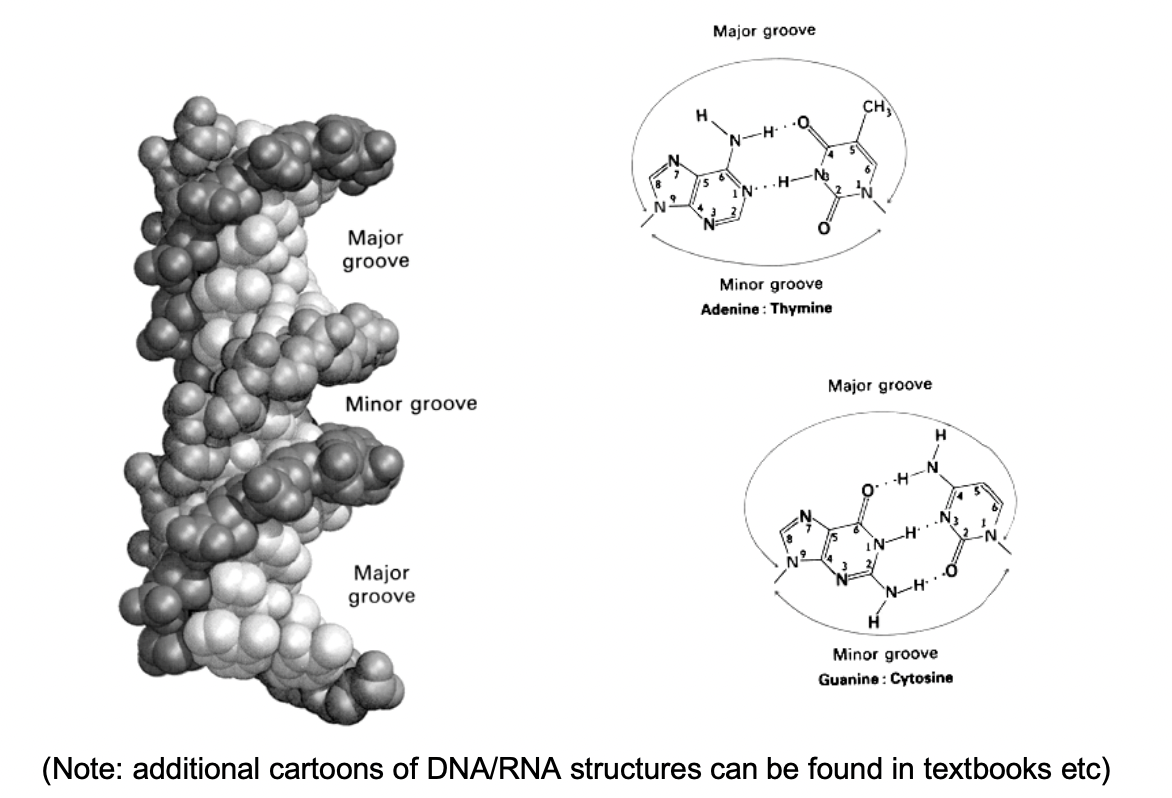
(Role of grooves) Access to epigenetic information is via
base modifications accessible in grooves
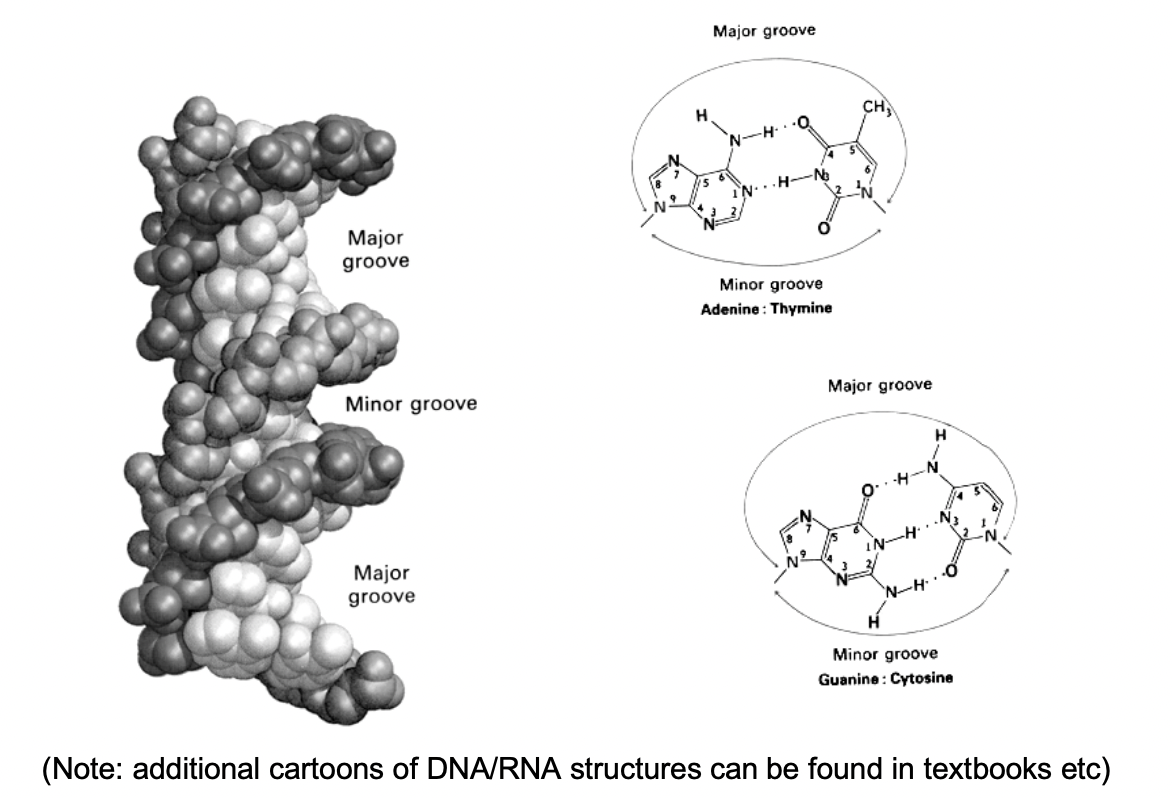
Interaction of DNA with sequence-independent binding proteins (e.g histones) also achieve through
interaction with the backbone
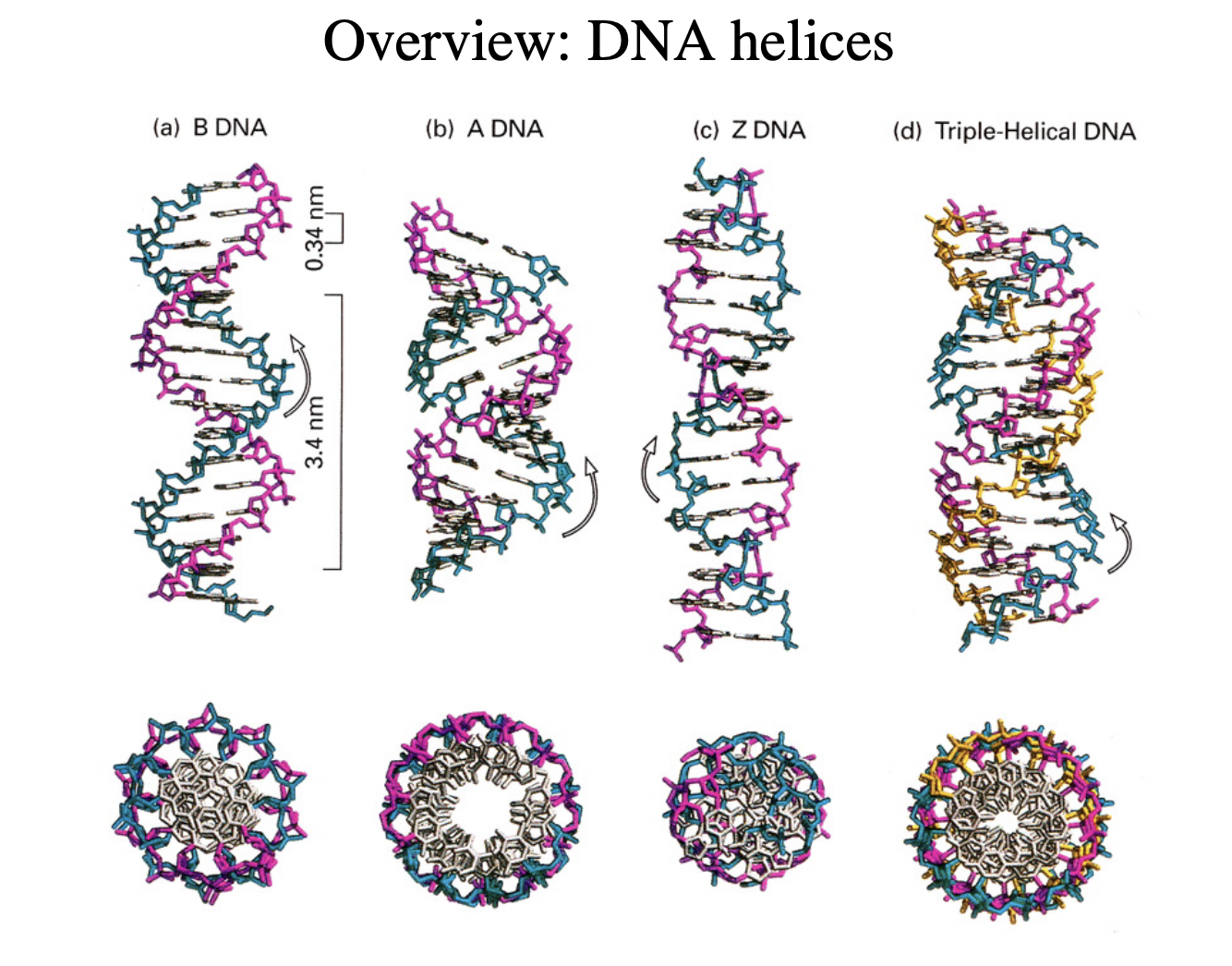
Types of forms of DNA
B-form→ right handed double helix
A-form→ right handed
more open
major/minor grooves not as distinct
seen more in RNA
Triple helix
when B strand has added things to it
Z DNA
left-handed
GCGC etc flipped so different direction
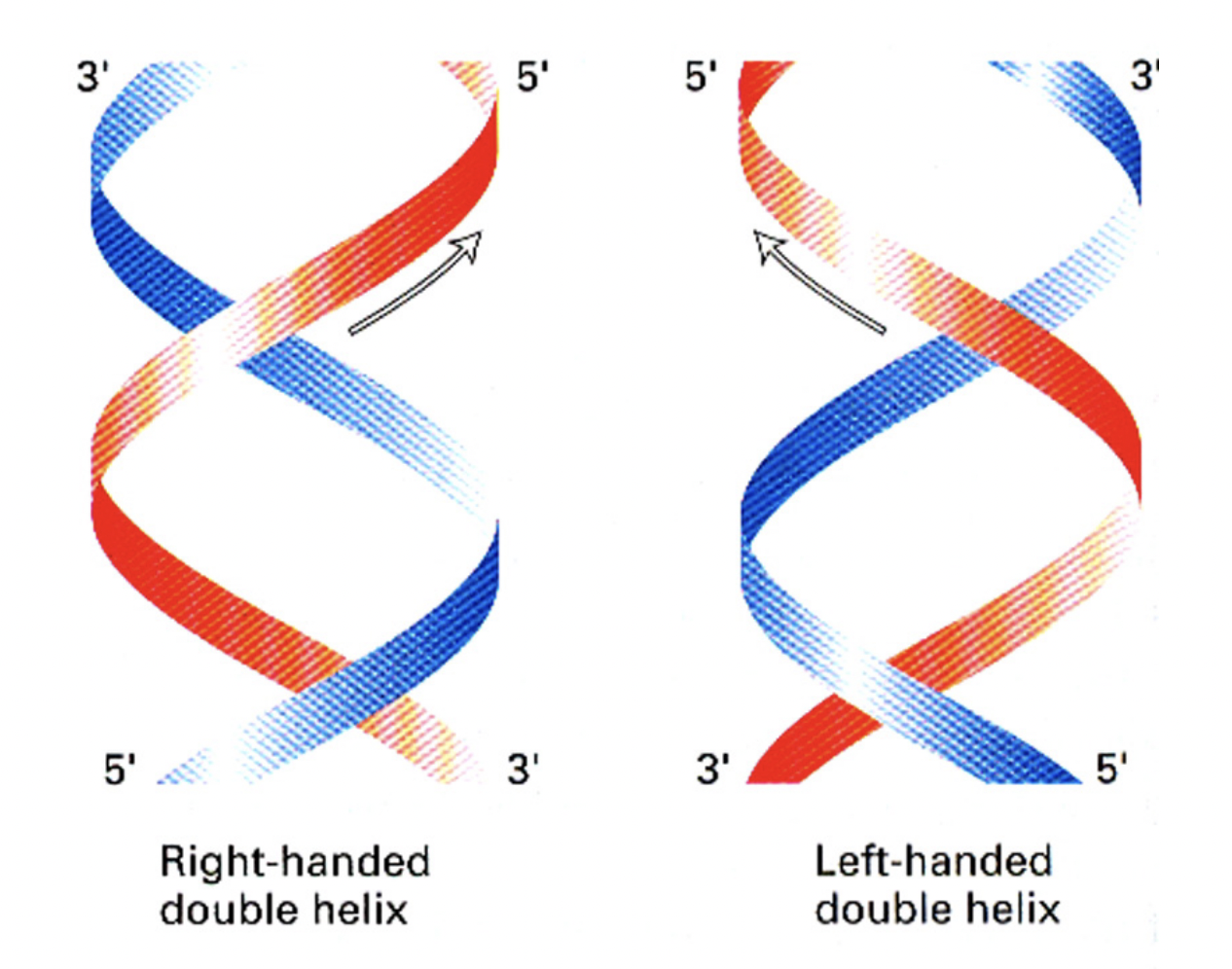
Right handed double helix vs Left handed double helix
to do with moving perpendiuclar finger hand up either arm thing?
How can DNA topology be visualied
electron microscope
Small circular genomes of eurkayotic DNA viruses can be seen in 2 configurations
relaxed
supercoiled
e.g SV40, polyoma
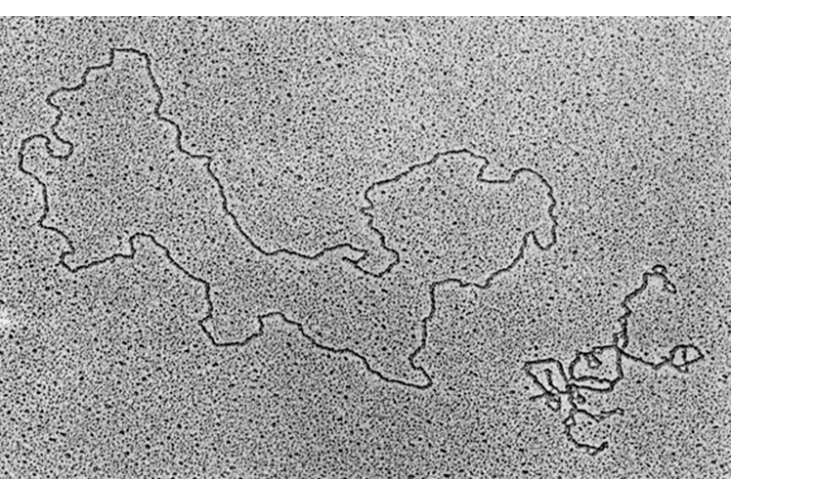
Relaxed
forms open relaxed ring
Supercoiled circle
Helix coiled around itself
forms more compact strucutre (compared to open relaxed)
Use of supercoiling in bacteria
to fit DNA into small cell
incontrast with…
Use of supercoiling in eurkaotic circular DNA
consequence of its association with histone proteins
to form nucleosomes
Supercoiled→ relaxed
by DNA Topoisomerase enzymes
transient breaking and resealing of the DNA backbone
DNA modifications usually to which base
cytosine
but can also be guanine
How cytosine modified
meythlation into DNMTs
oxygenated into TET
coniuted oxygenation
becomes more and more bulky
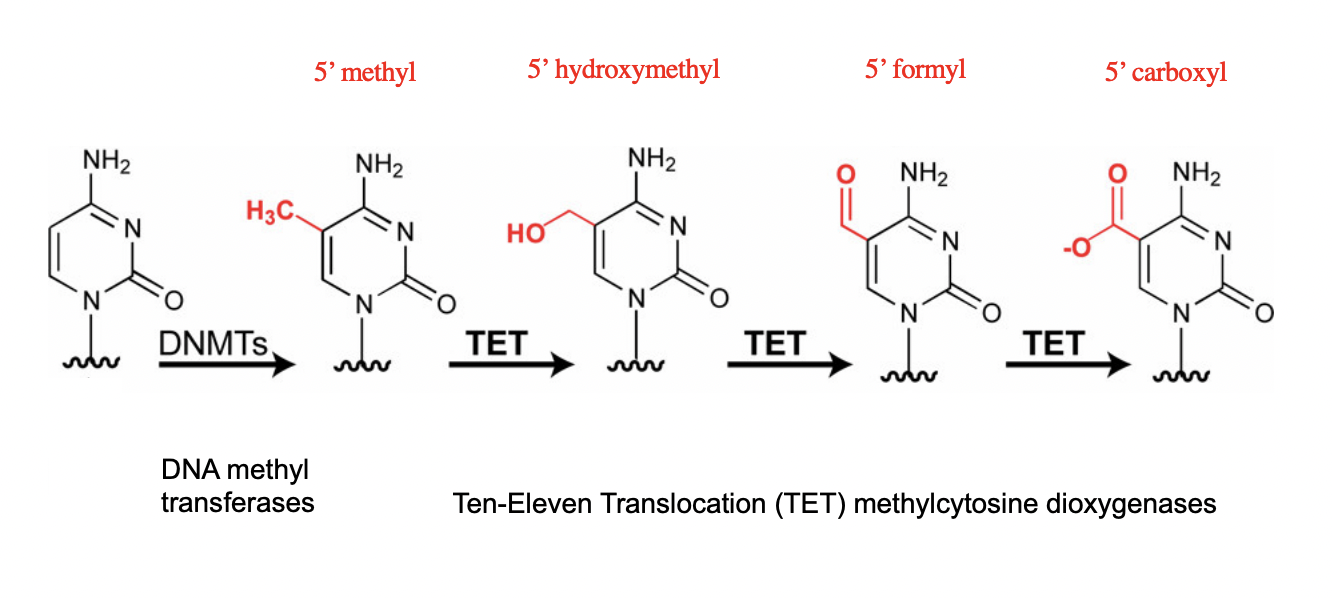
What does this result in?
Cytosine sticks out
epigenetic effect
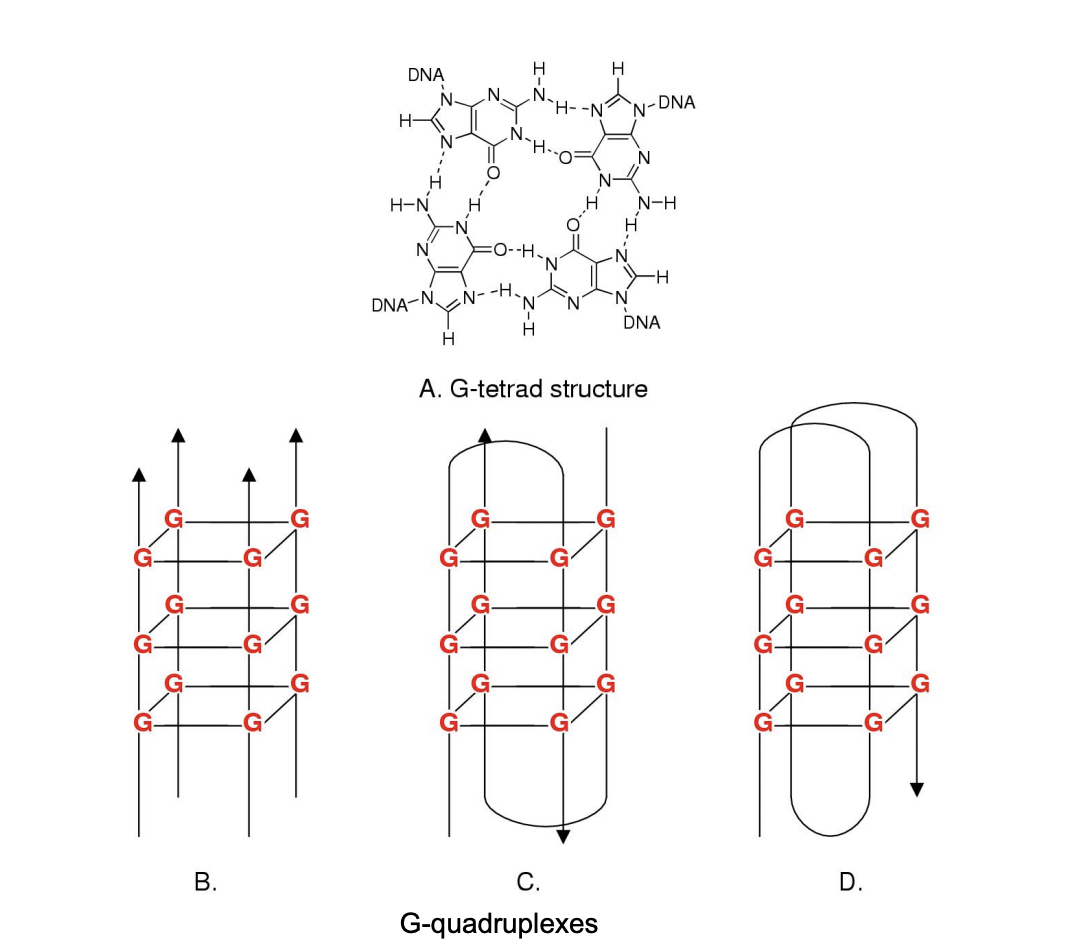
Guanosine modifications can form
secondary strucutures:
stable tetrad strucutre
in test tube and in cells
form higher order strucutres (see image)
RESULT: plastic DNA modifications
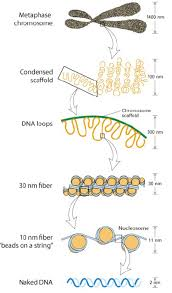
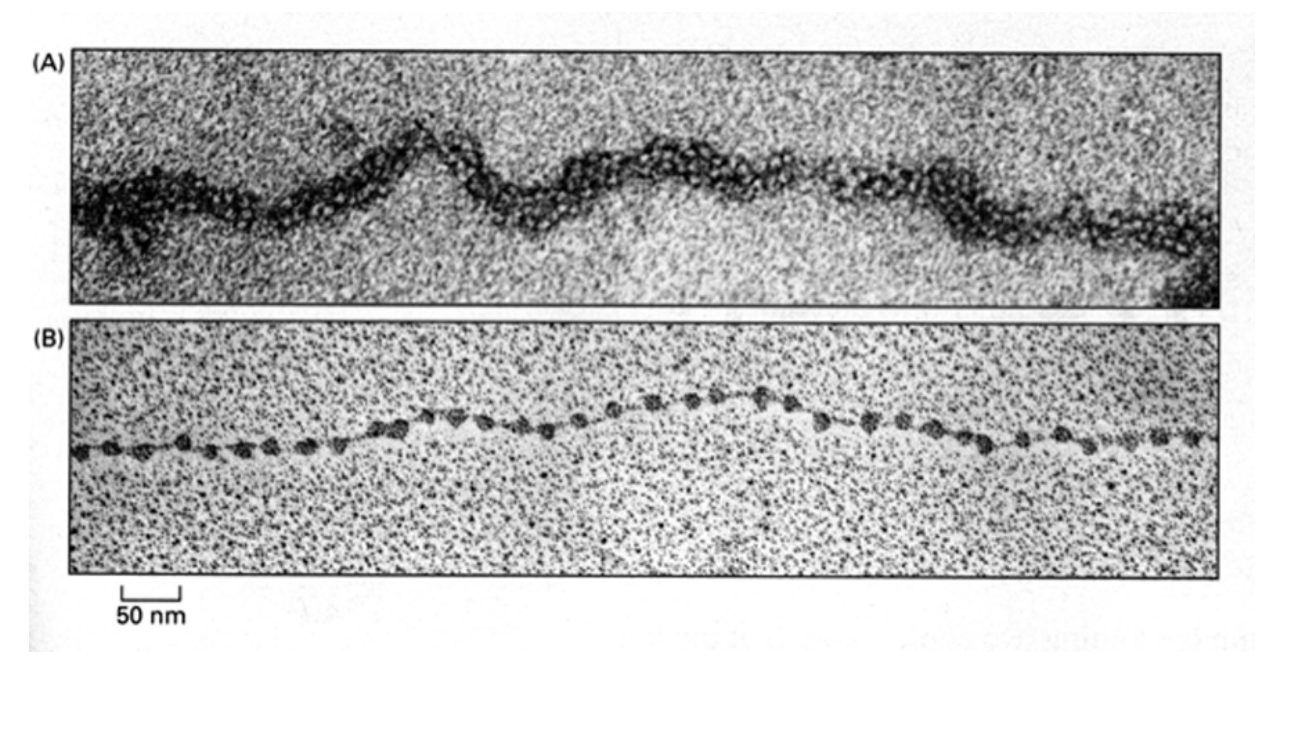
What Electromircopy has shown about nucelosomes
regular arrays of nucleosomes
along the genomic DNA→ 1/100 mm in diameter
→ constituting a chromatin fibre
beads on a string
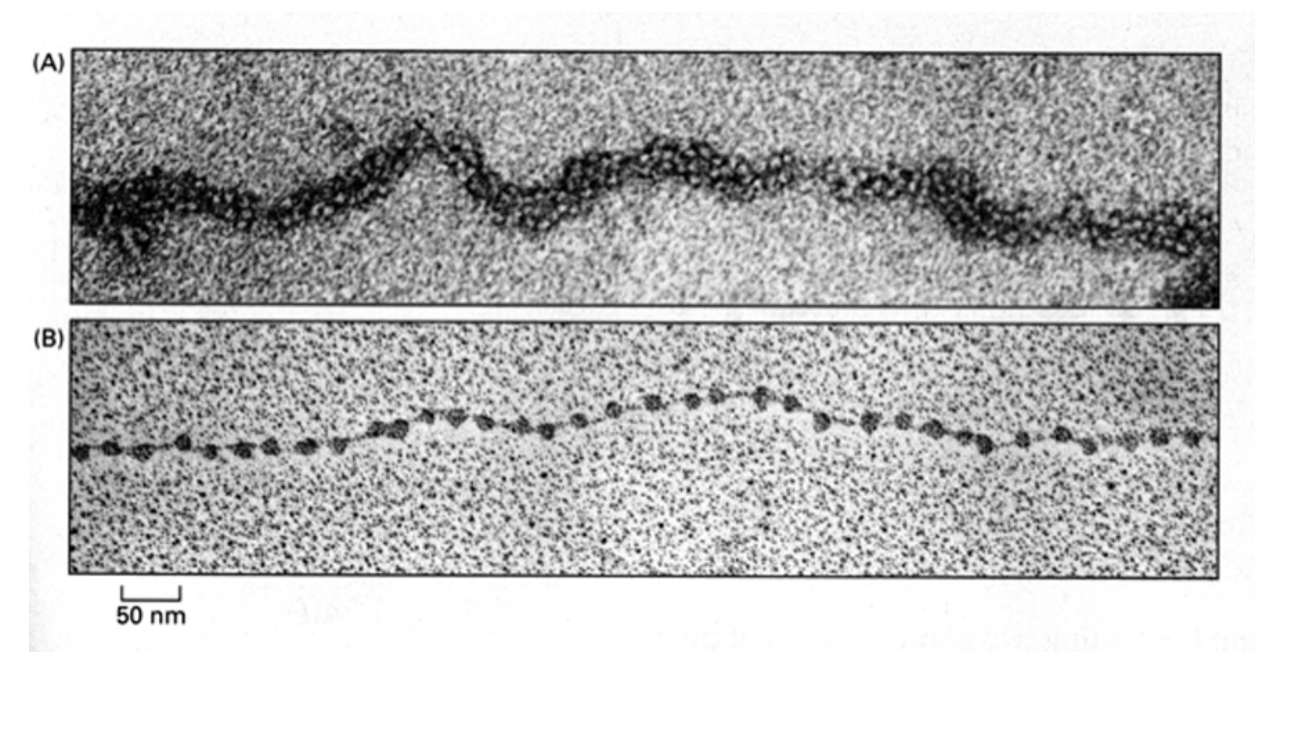
30 micrometres physiology conditions
10micrometres regular pattern
DNA linker shown
nucleosome blobs
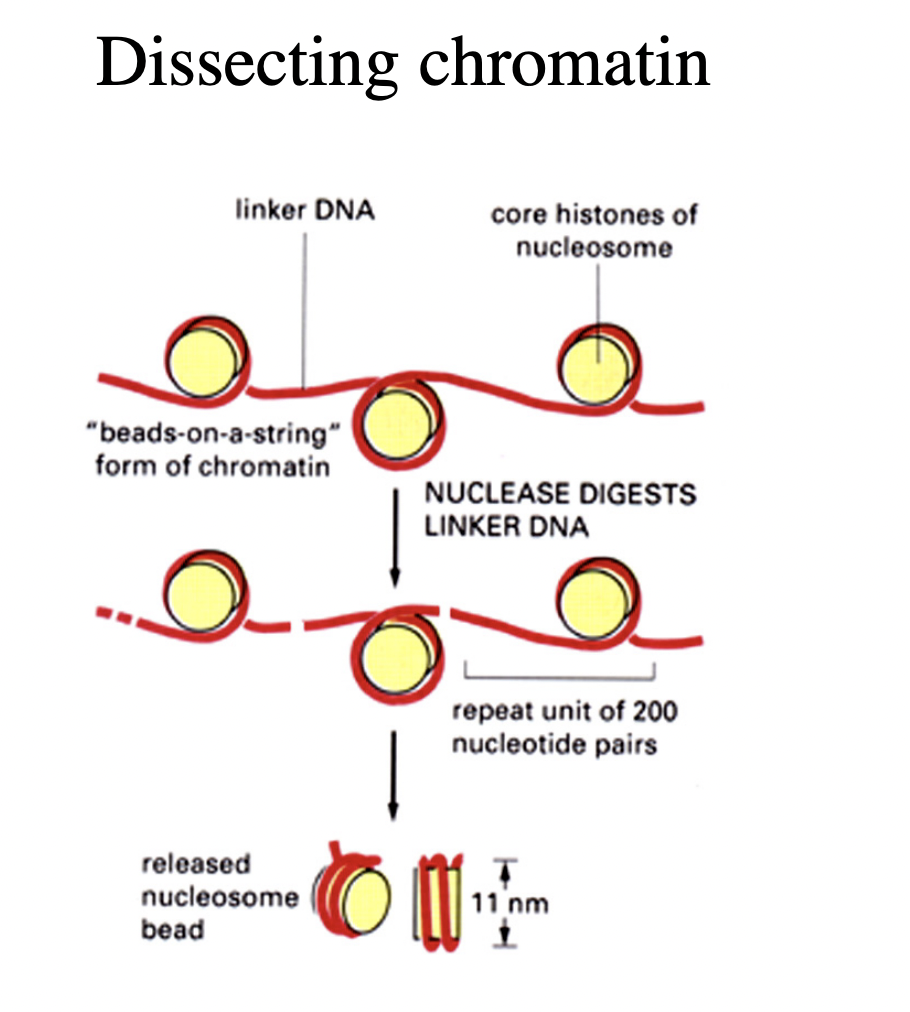
What do certain DNA endonucleases do to DNA
e.g Micrococcal Nuclease (only cuts linker DNA)
release nucleosomes as discrete substructures
containing:
fragments of genomic DNA
strucutrual histone proteins
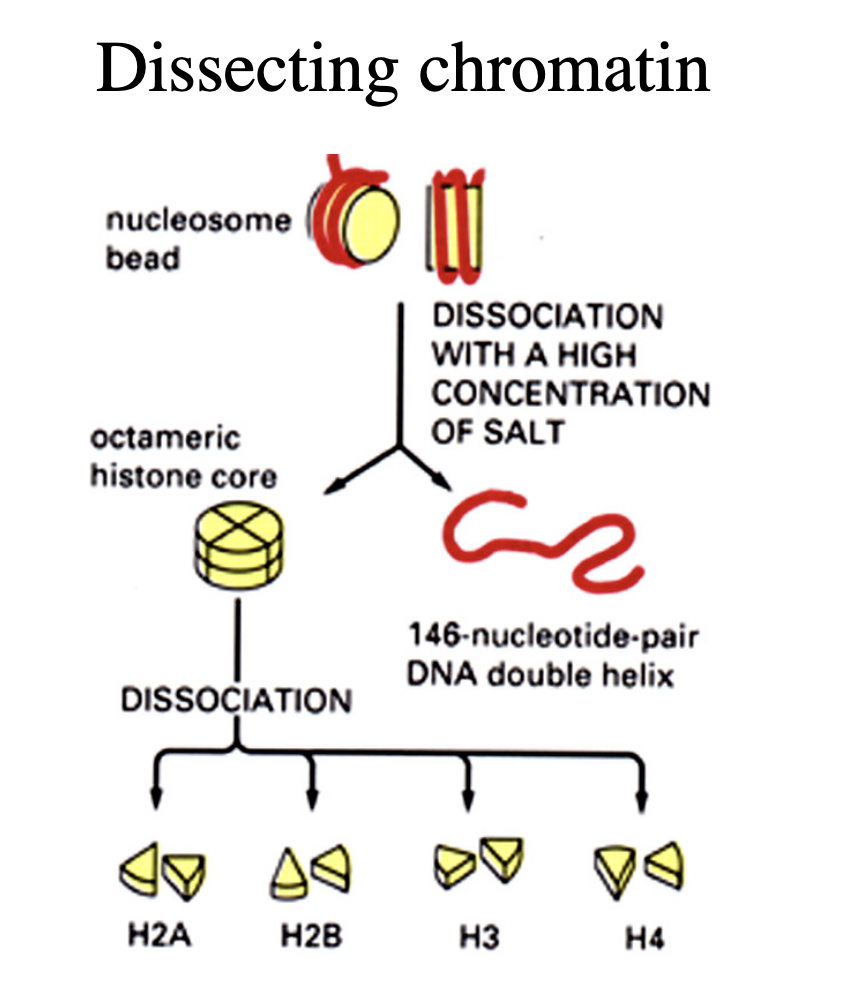
How can the DNA and histones be separated further?
high salt treatment
or
denaturation
Although the core is 146 bp, further digestion with DNAse 1 shows…
regular periodic patern of 10-12bp fragments
i.e number of base pairs per full helical turn
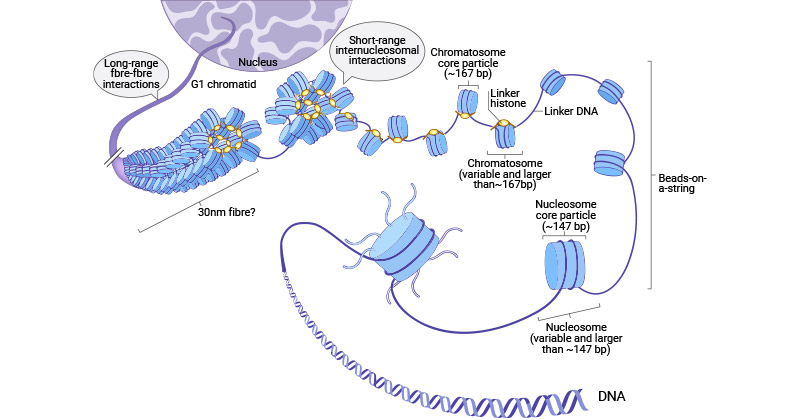
What does this indicate NEED TO REVIEW THIS CLEARER LOOK AT BoC NOTES
the DNA is wrapped around the surface of the histone core octamer
it is around the outside surface of the histone
not surrounded by the histone
this is beecause we got regular strands from this digestion
Digest the DNA with histone attached and see wht DNA is available→ with further digestion, will be on surface if there
we get regular stands which means that it is turned 2 ½ times around the histone
so must be on the surface
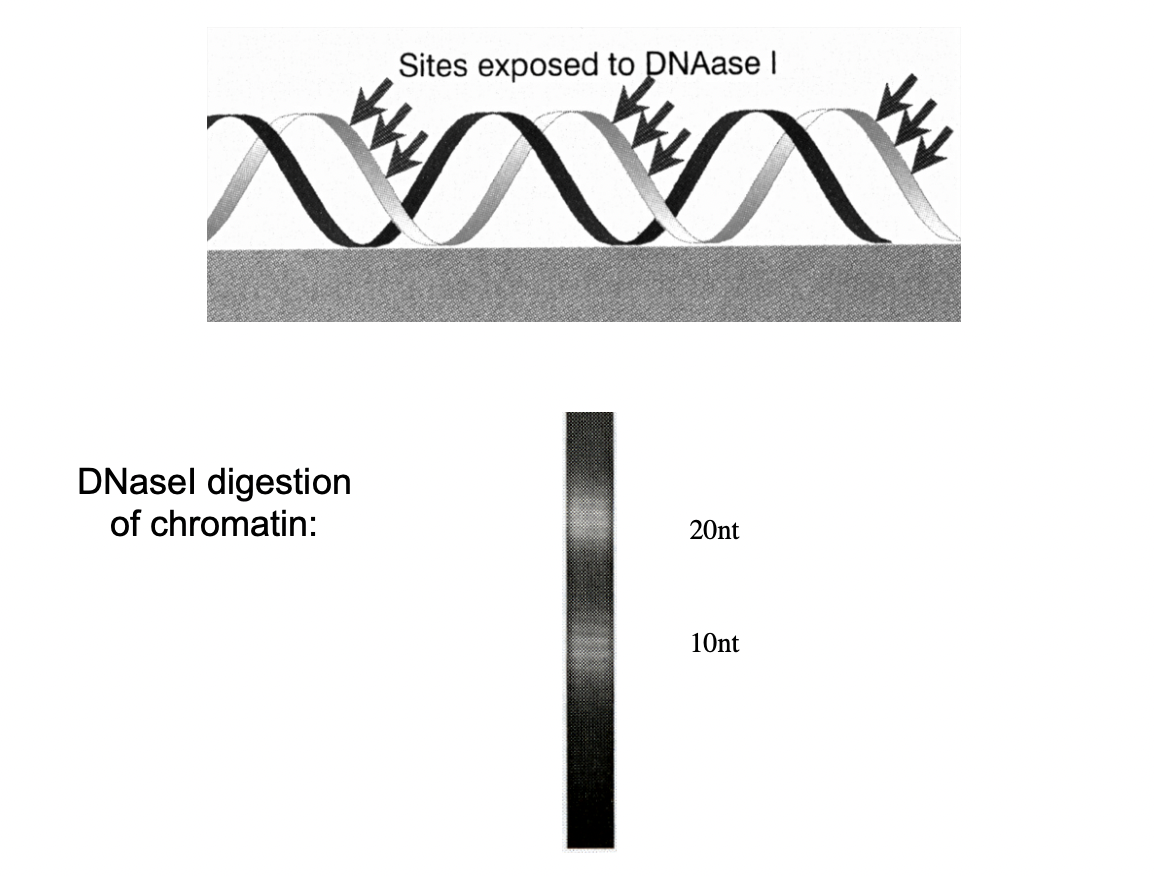
The nucleosomal wrapping of DNA around the histone core leads to…
the formation of one contrained supercoil per nucleosome
→ this is released when histones are removed
wrapped around twice
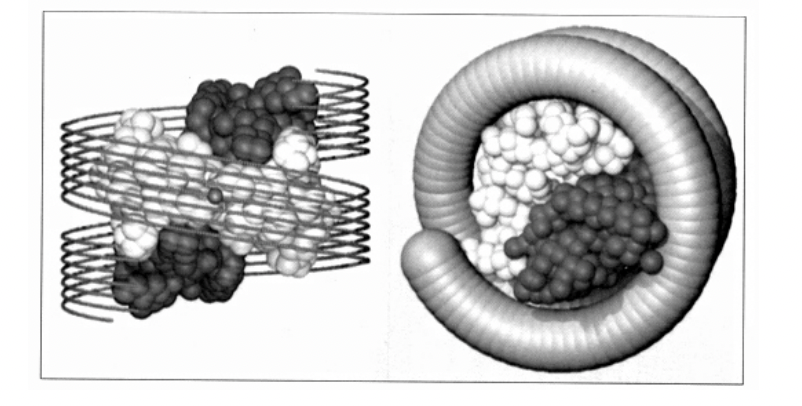
HIgher resolution of nucleosome core particles→ derived from cystallography and X-ray diffraction
front and side view
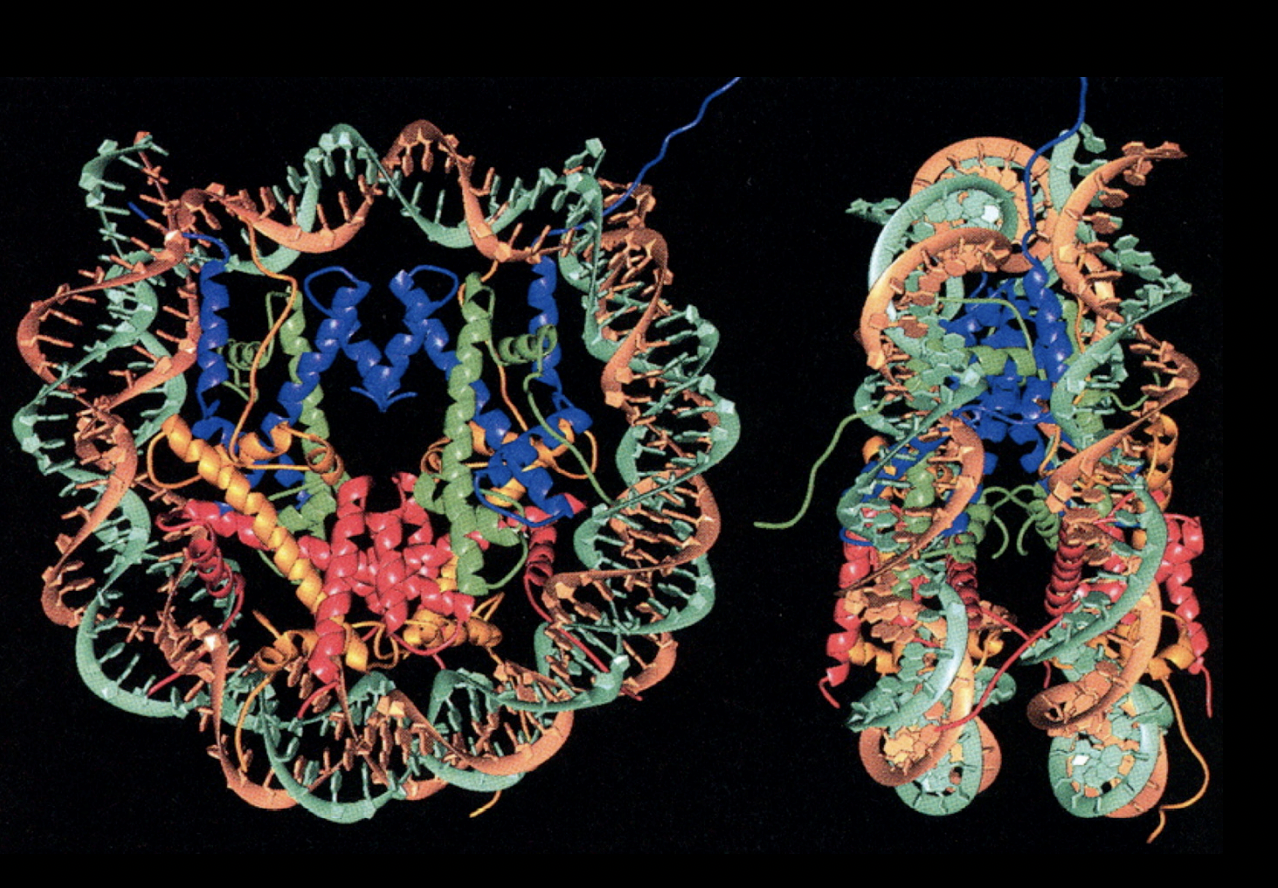
The histone unfolded
shows the components
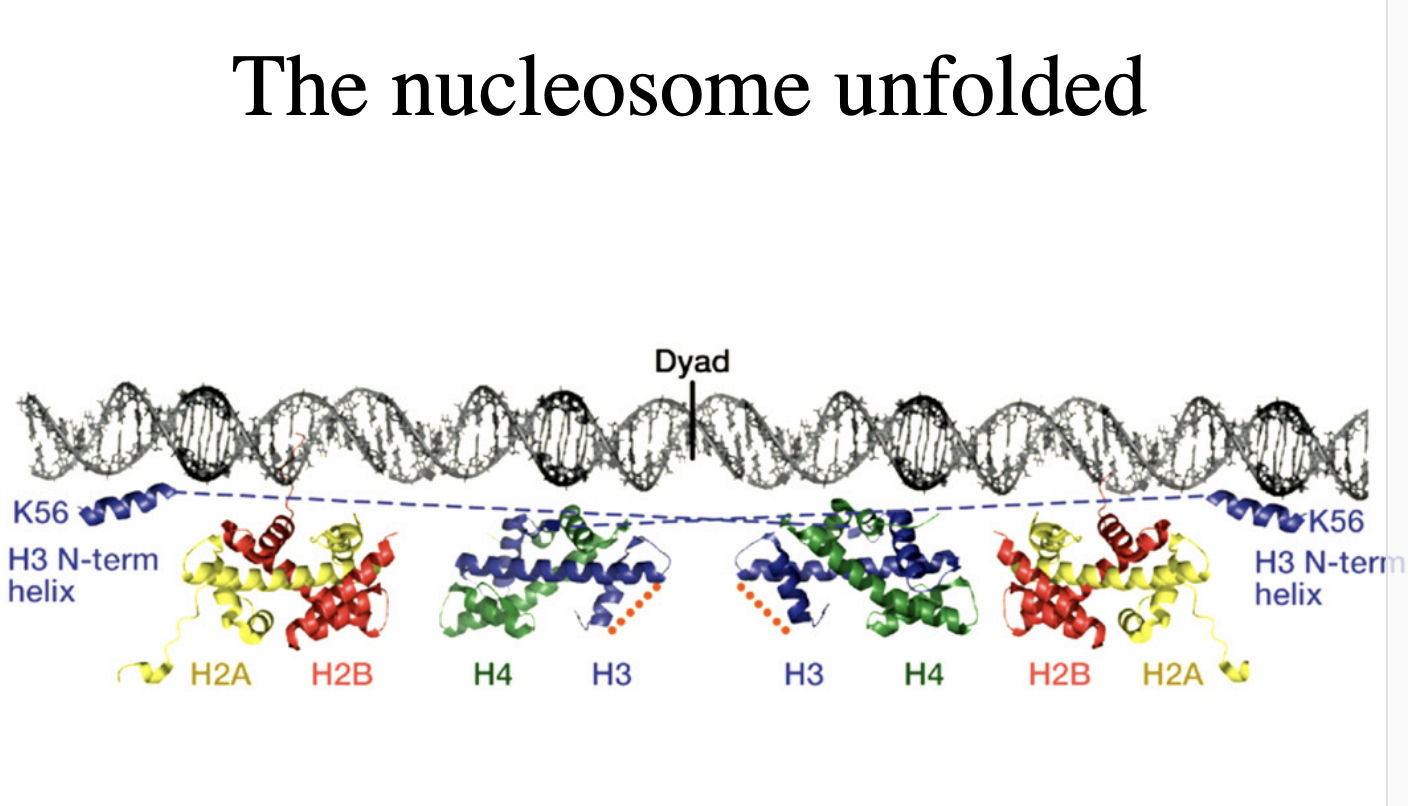
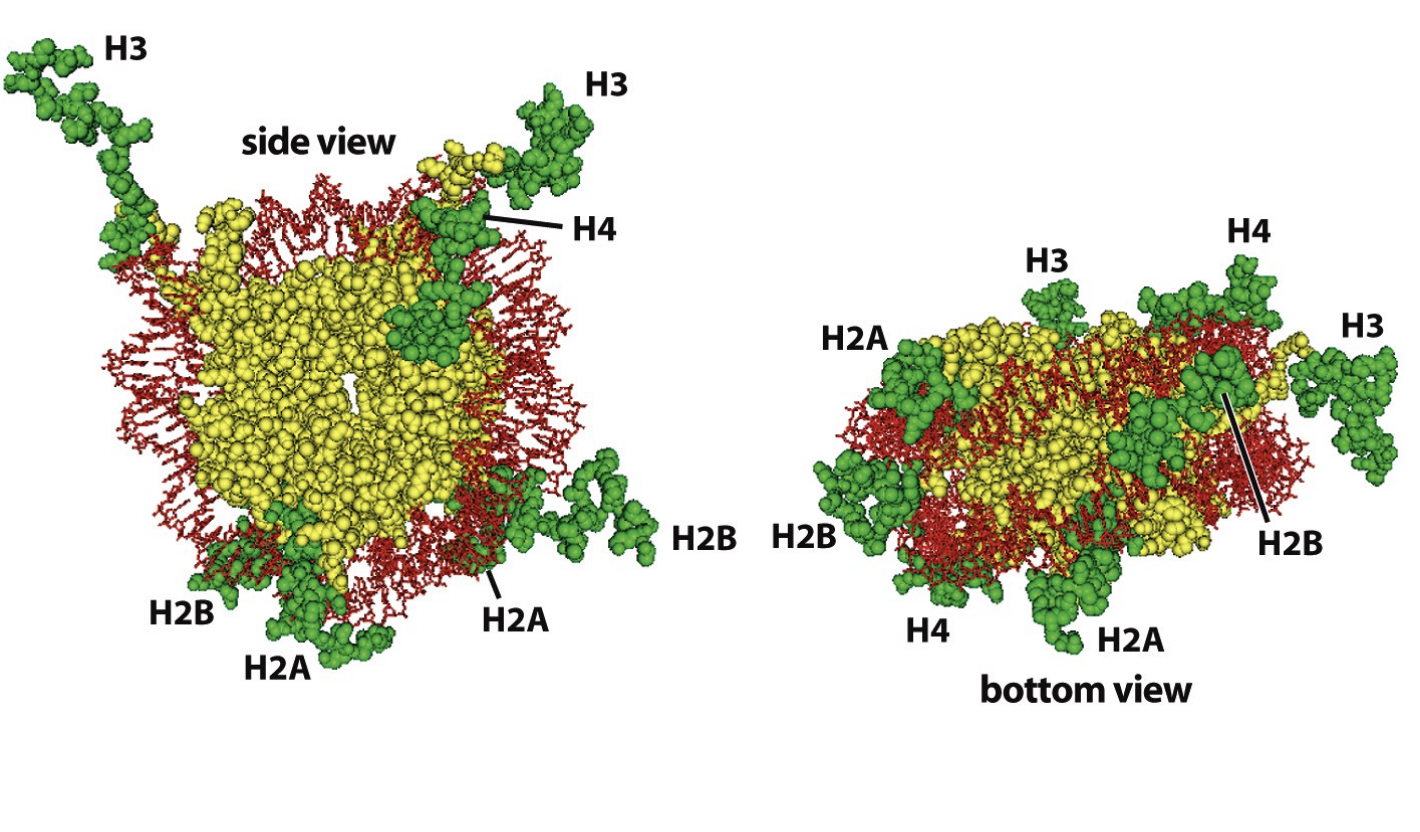
Nucleosomes core particles (in all eukarotic organisms)
146 bp of DNA
octameric histone core:
with two copies of each core histone
(H2A, H2B, H3, H4)
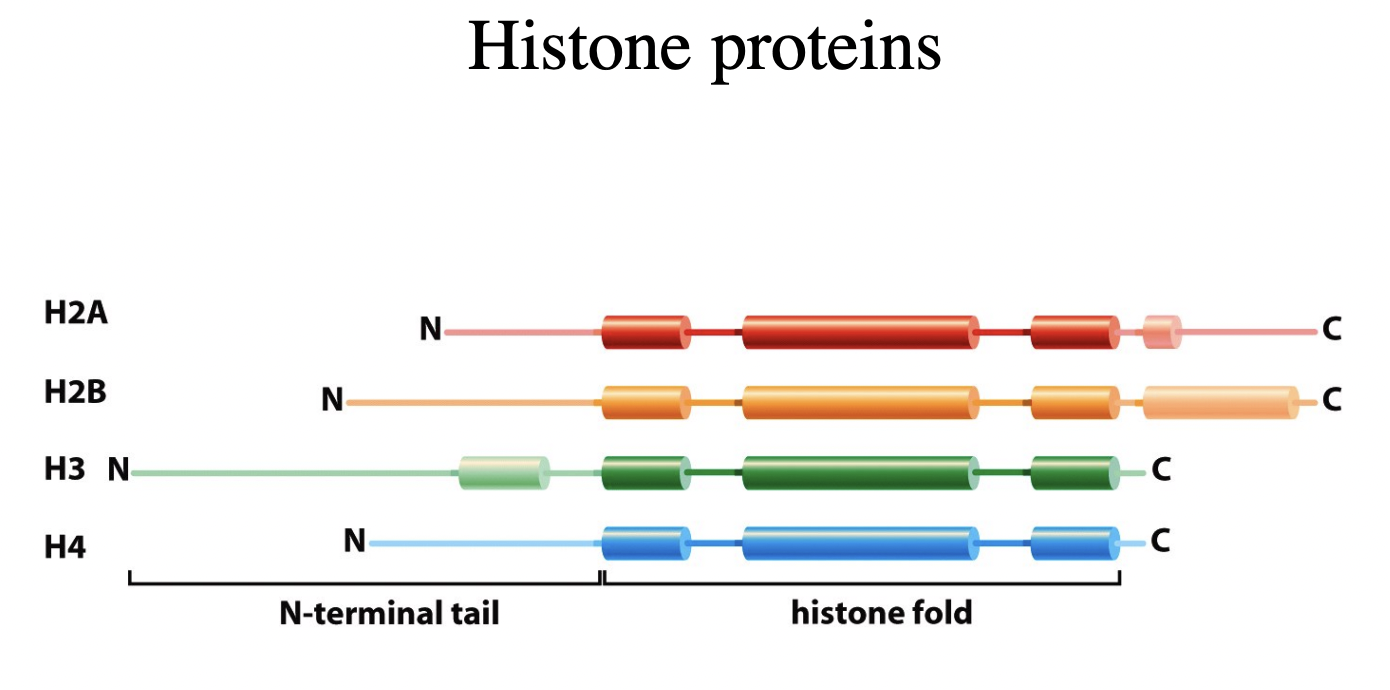
How do histone proteins form stable substrucutures
Heterotetramers of two molecules of each:
H3 and H4
Heterodimers of
H2A and H2B
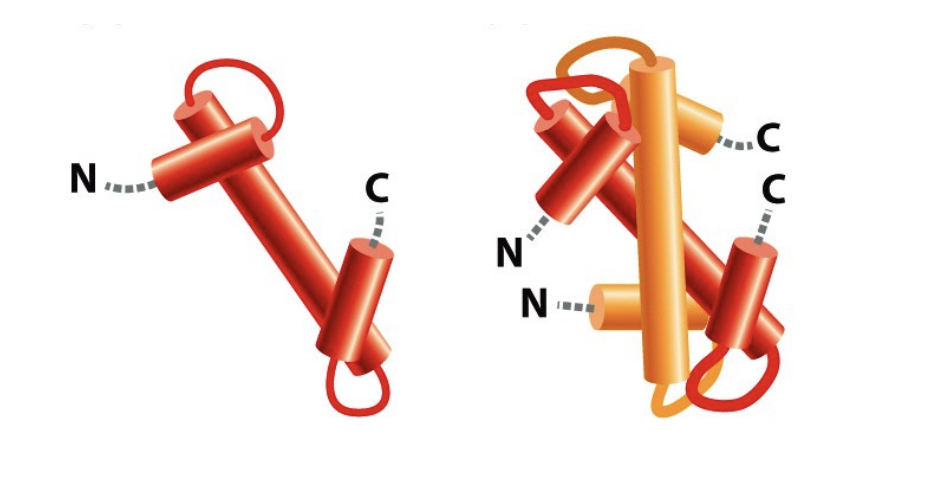
The histone protein folds the handshake motif (can do with hands)
image shows an H2A and H2B handshake motif
These heterotetramers then form the histone octamer, how
one tetramer
two dimers
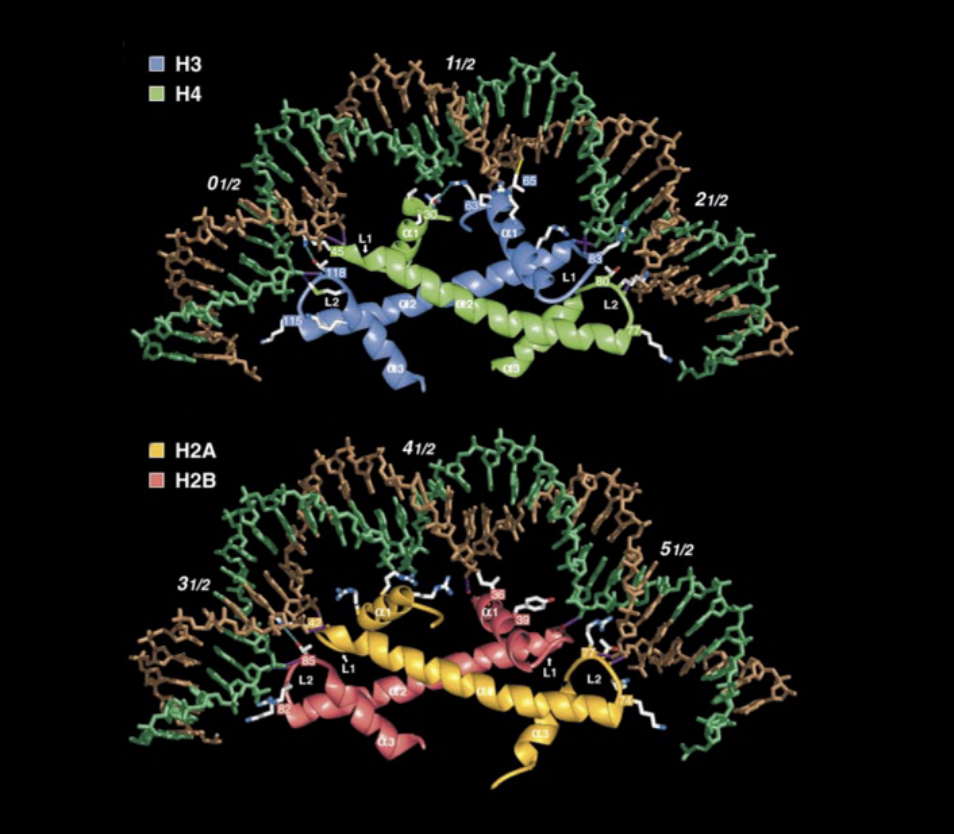
Role of histone octamer in nucleosome
forms the strucutrual core
strucutre of nucleosome has been resolved at atomic resolution by
X-ray diffraction of pure nucleosome crystals
high-resolution strucuture (image)
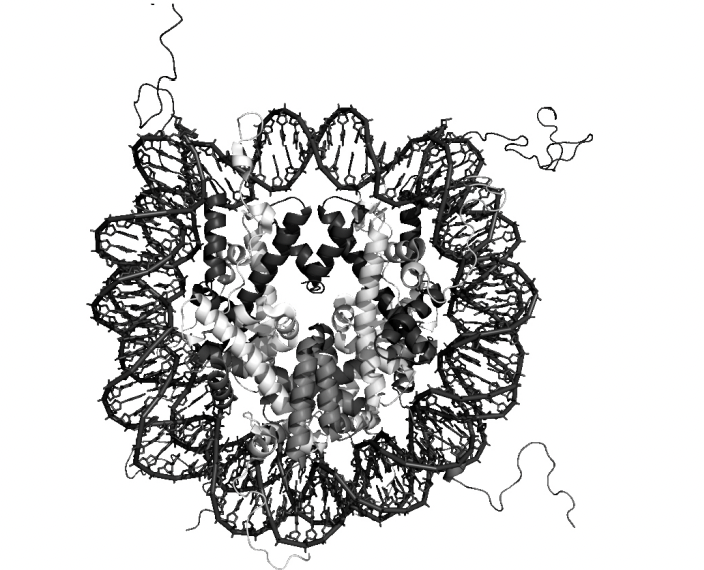
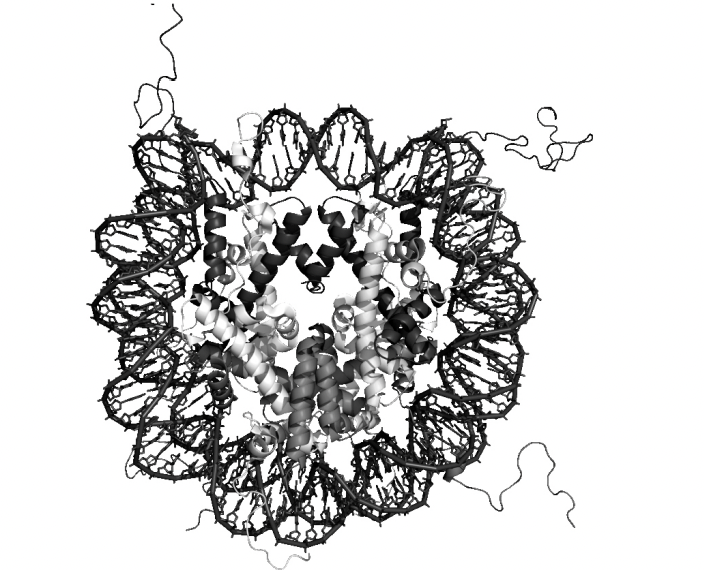
Details of this strucuture
central glocular domains of histones proteins are located on inside
N and C terminal tails of histones extend out from the core strucuture
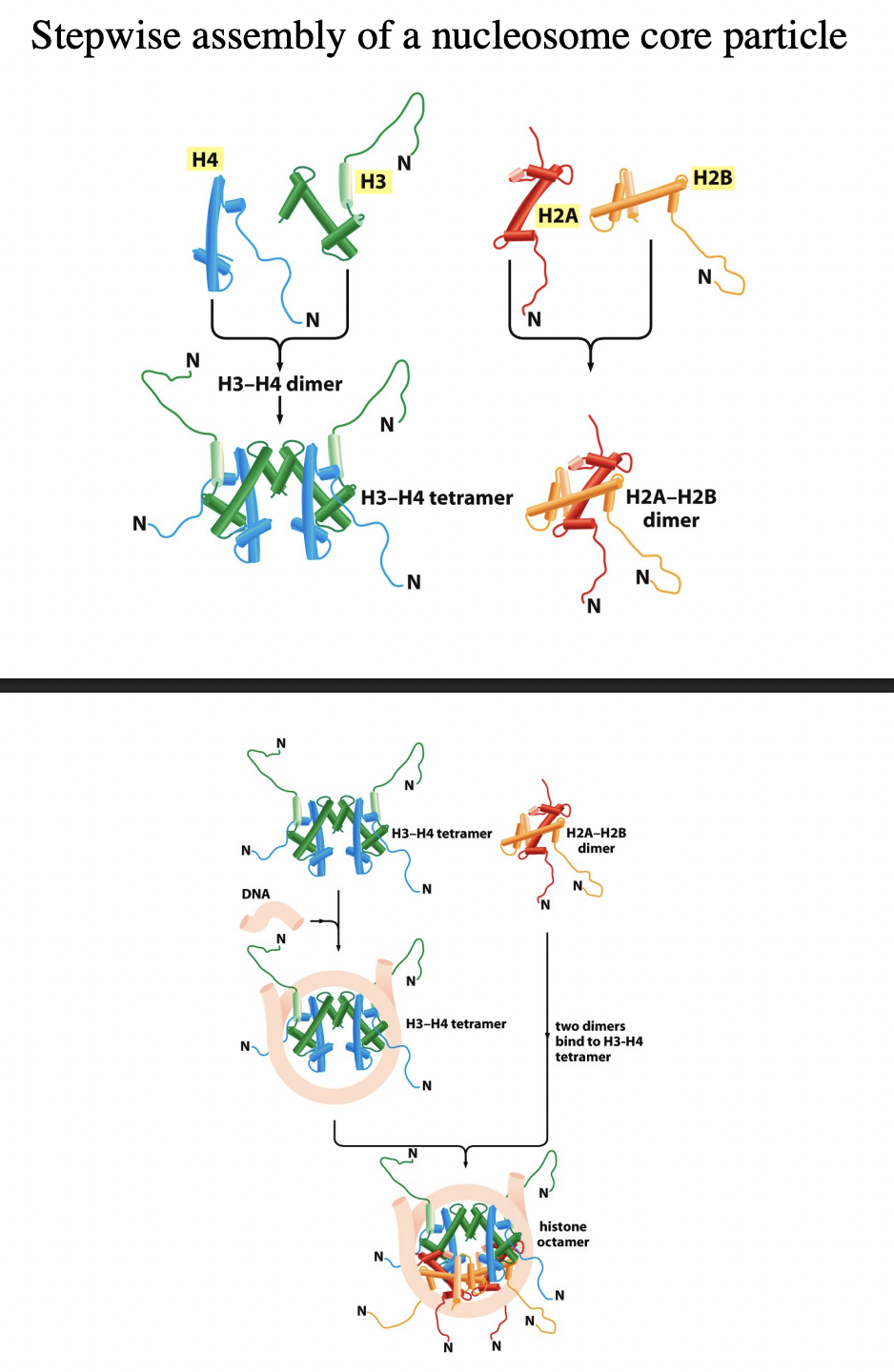
How are nucleosomes assembled from histone subcomplexes? Stepwide assembly
tetramer of (H3-H4)2 contacts DNA
this is subsequently joined by two H2A-H2B dimers
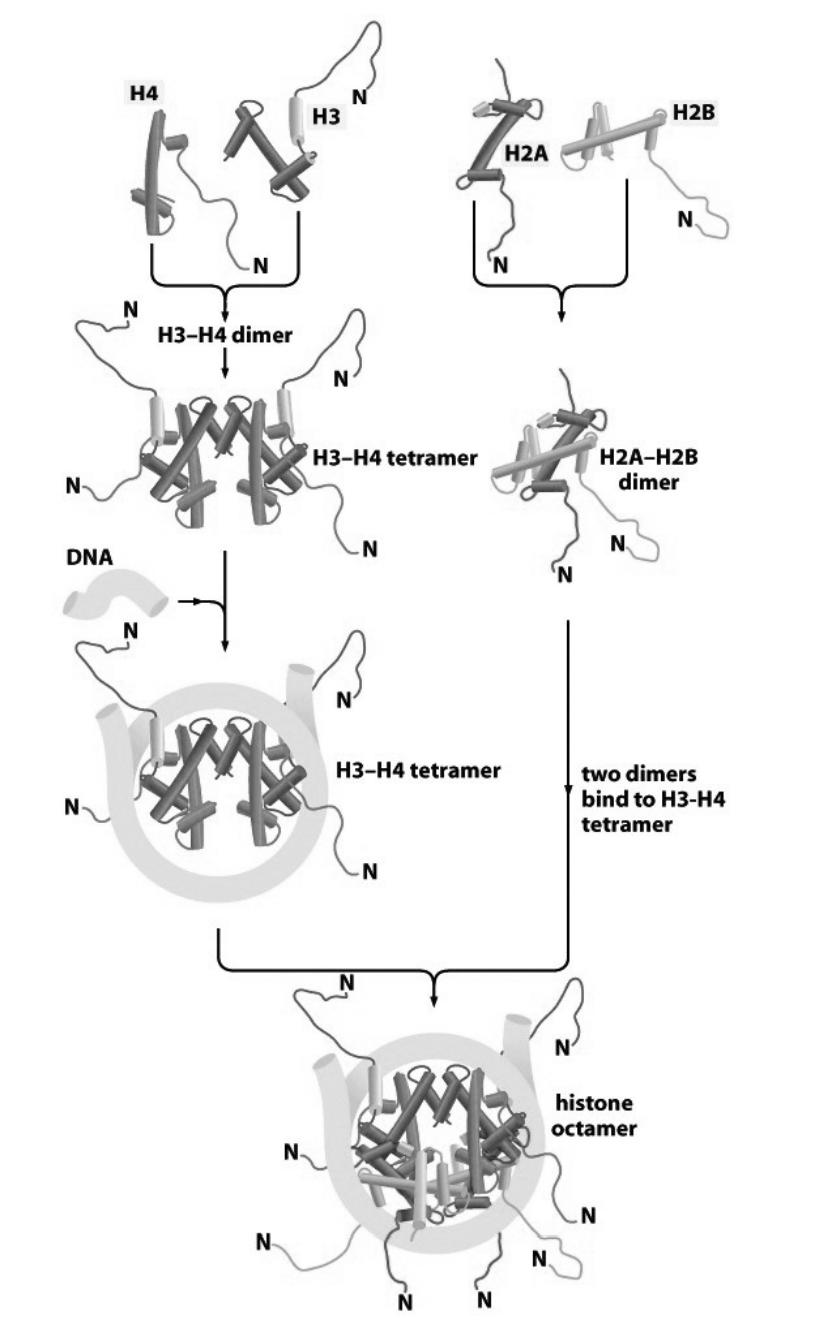
conditions for when this reaction occur?
purified components
in vitro
if the salt conditions are carefully controlled
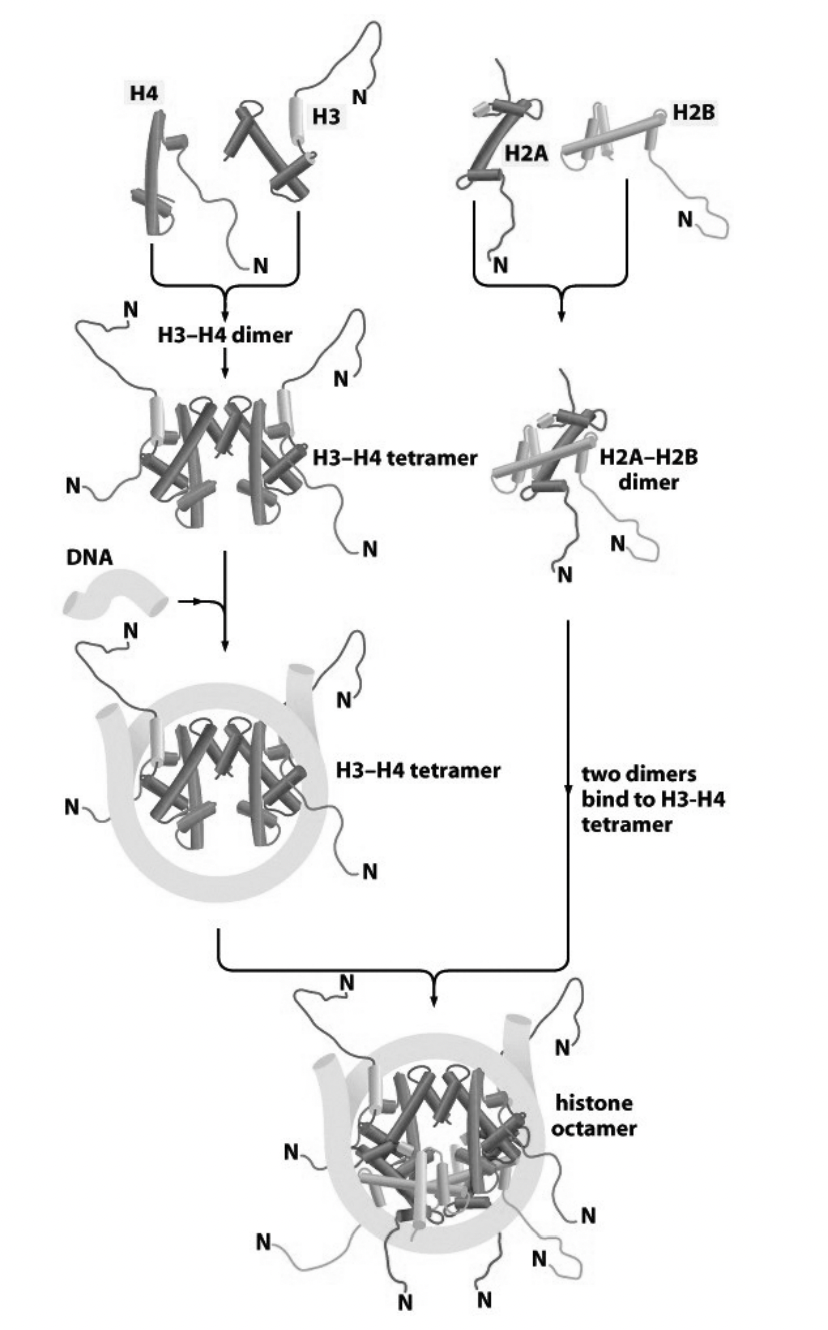
What about in vivo?
mediated by:
histone chaperones
chromatin assembly factors
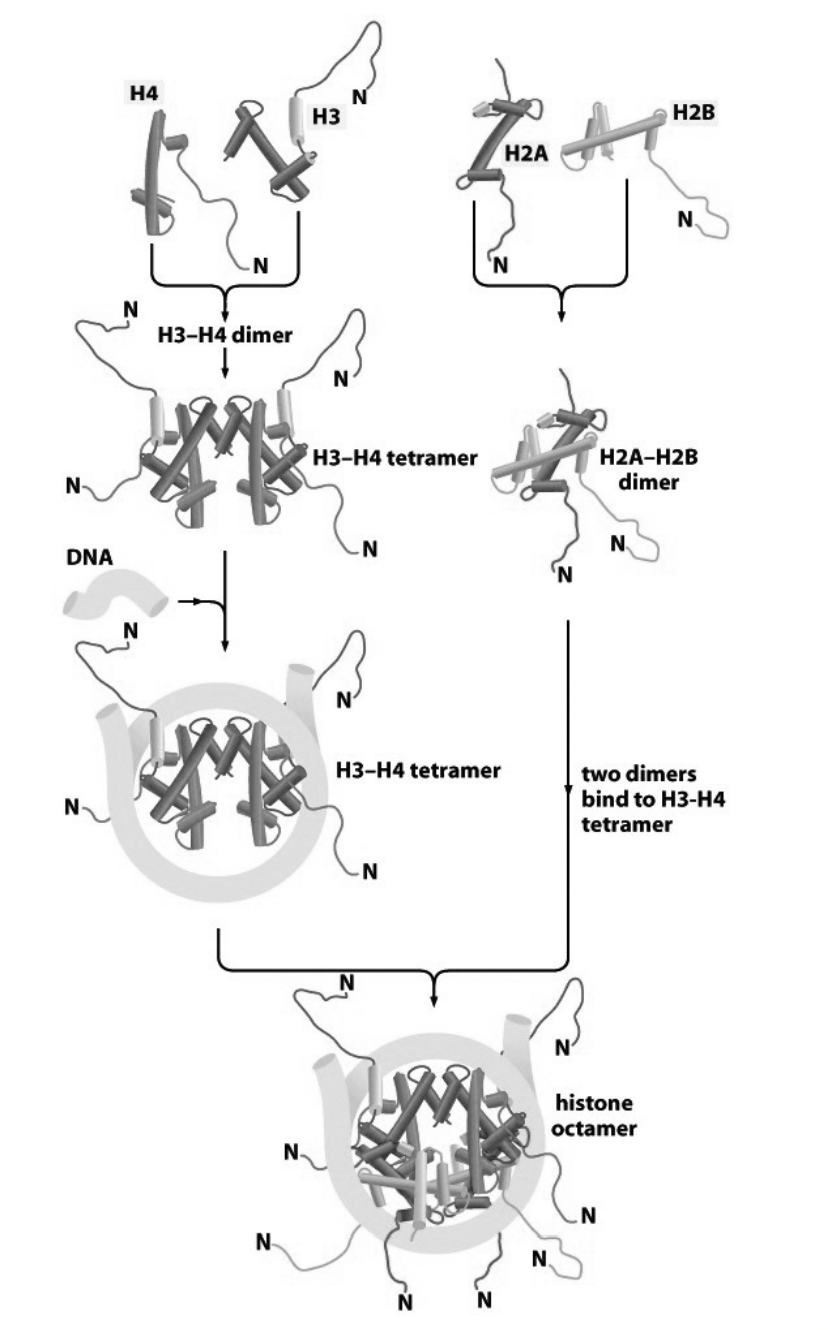
(more detailed Stepwise assembly)
Handshake motif od H2A and H2B (twise to make two dimers)
Handshake motif of H4 and H3 (twice to make two dimers)
H3-H4 dimers binds to make H3-H4 tetramer
This attracts the DNA and its force around tetramer (knuckeles and fingers are in contact)
molecular velcro
two H2A-H2B dimers bind
Histone octomer is made
NEXT adjacent nucleosomes connect with DNA linker
How are adjacent nucleosomes connected?
by linker DNA
Size of linker DNA?
varies between:
cell types
tissues
species
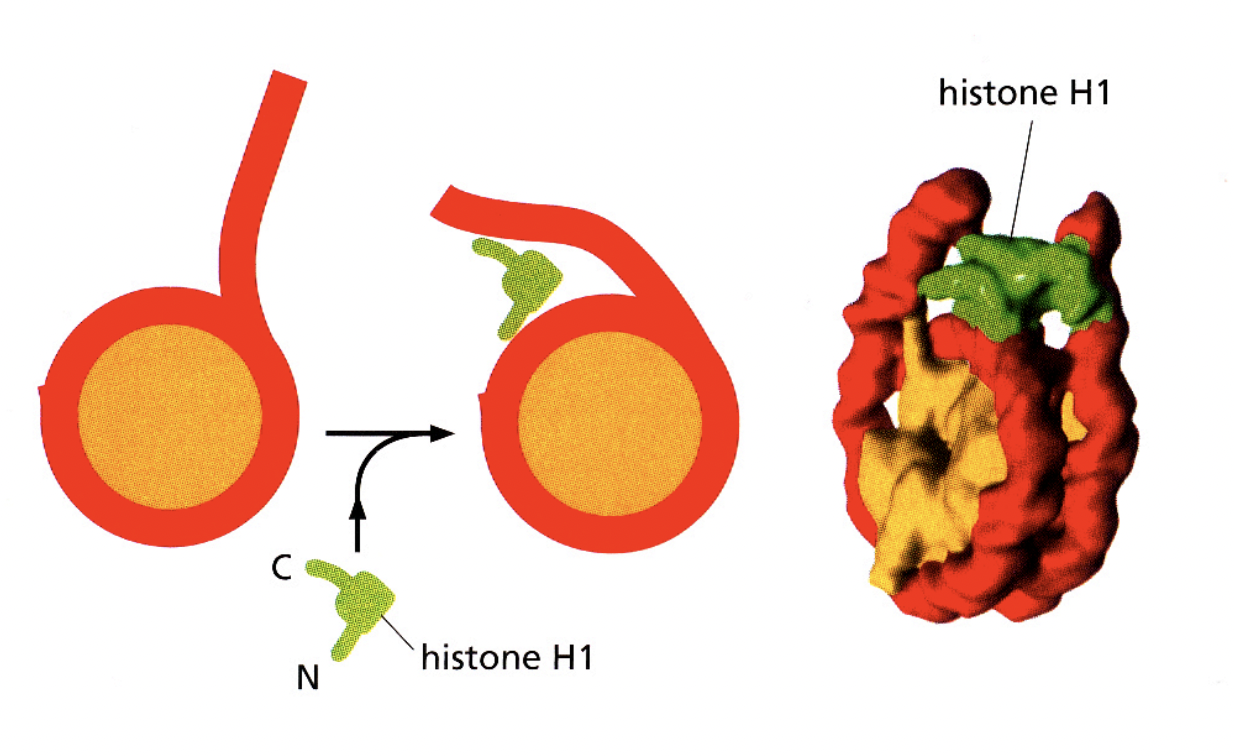
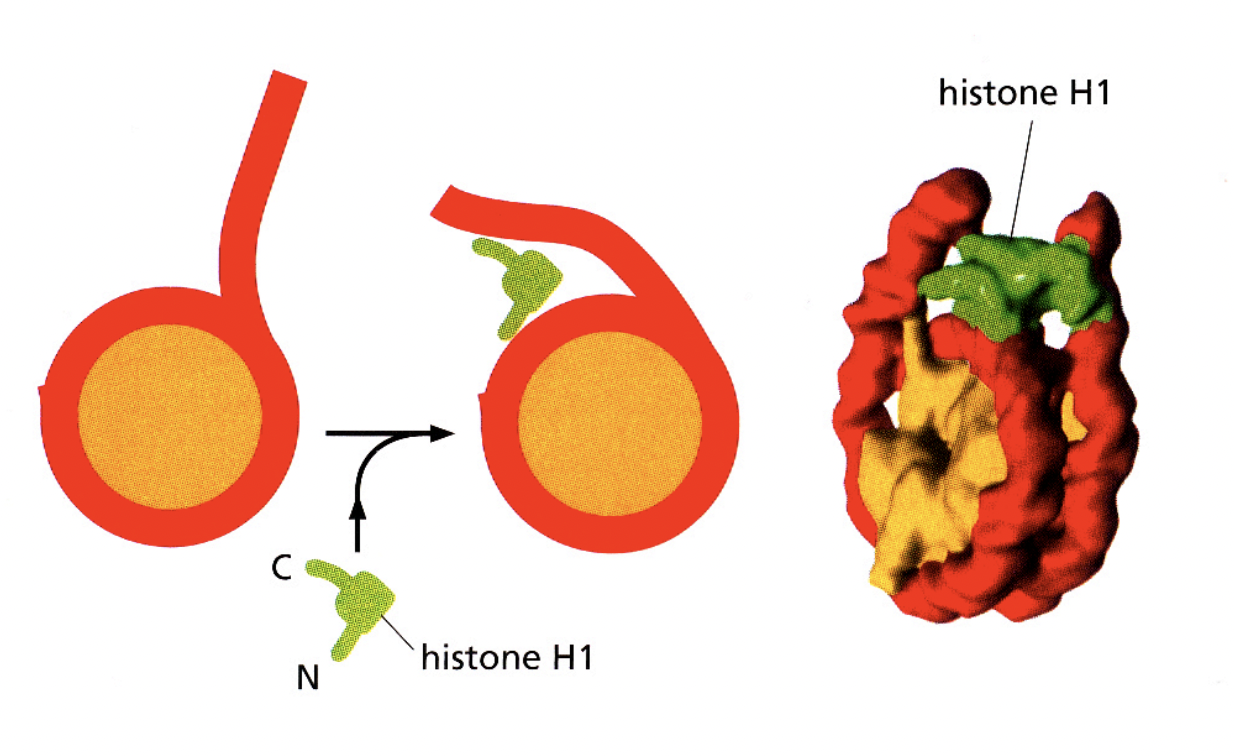
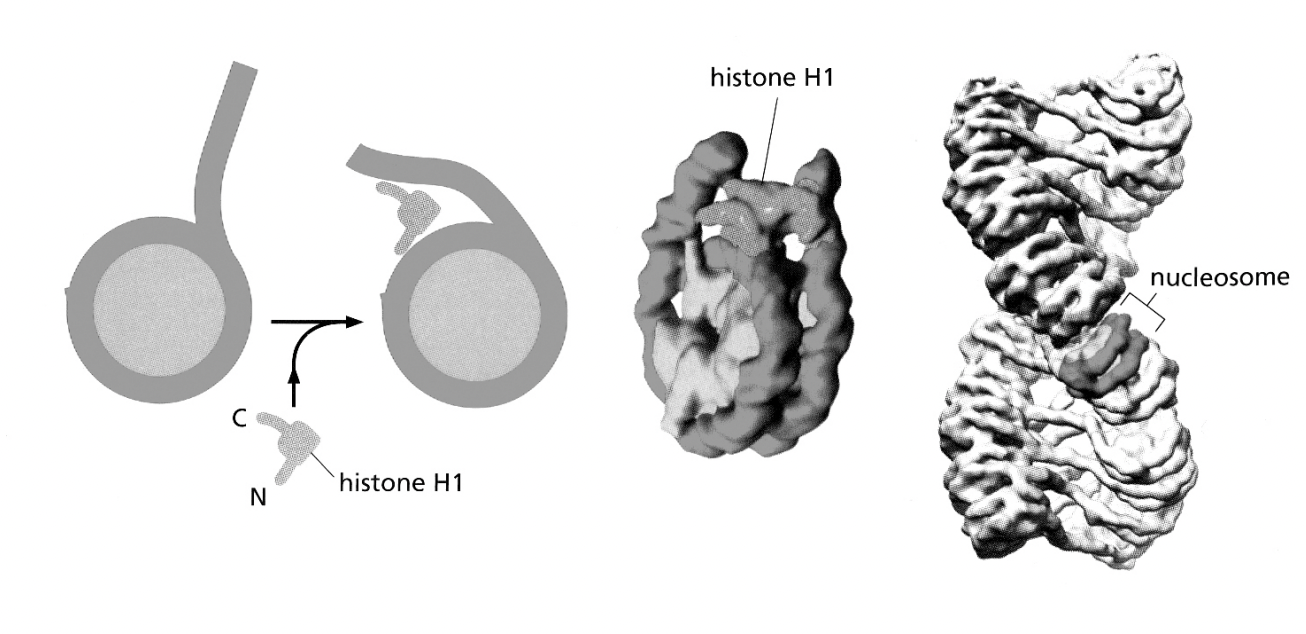
Associations of linker histone H1 with the linker DNA and nucleosome core are involved in forming…
higher order structures
H1→ helps to stabilise the higher order strucutures of the nucleosomes?
Modifications of histones, how?
tailes of the four core histones extend out from the nucleosome core
available to be acted upon
by protein modifying enzymes
Specific amino acids in the tails are often post-translationally modified by a wide variety of modifcations
Histones are proteins, and have N and C termini.
The N termini stick out a bit and so can be modified
e.g with acteyltransferase
What modifications can be done to thse amino acids?
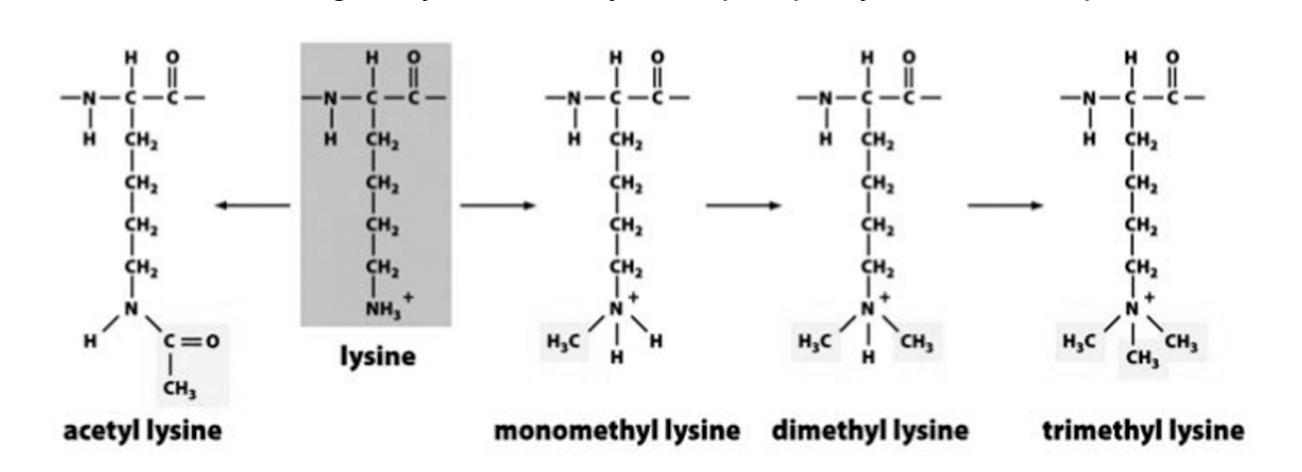
acetylation
methylation
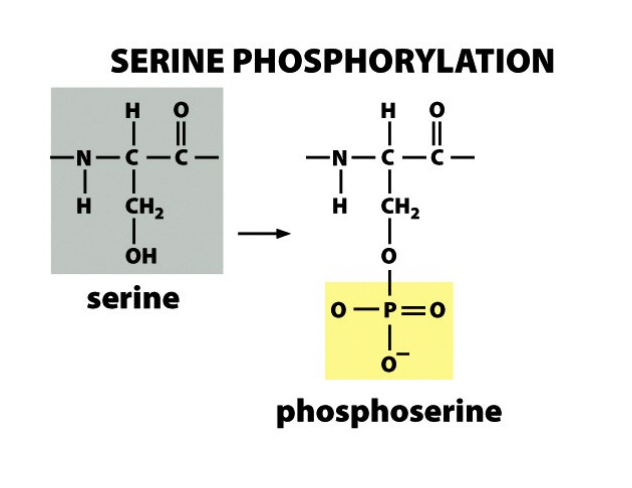
phoosphorylation
ubiquitination
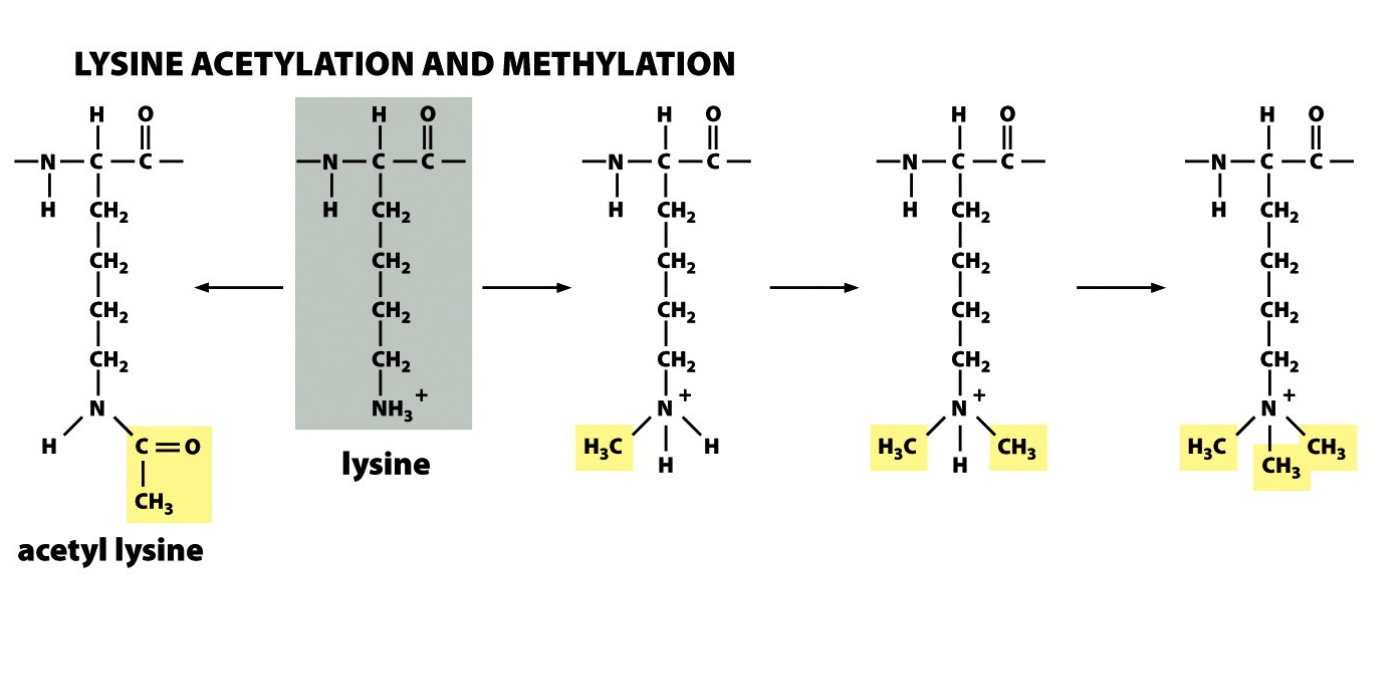
Lysine acetylation and methylation
Acetylation→ neutralise charge
Methylation→ fixed, and bulky
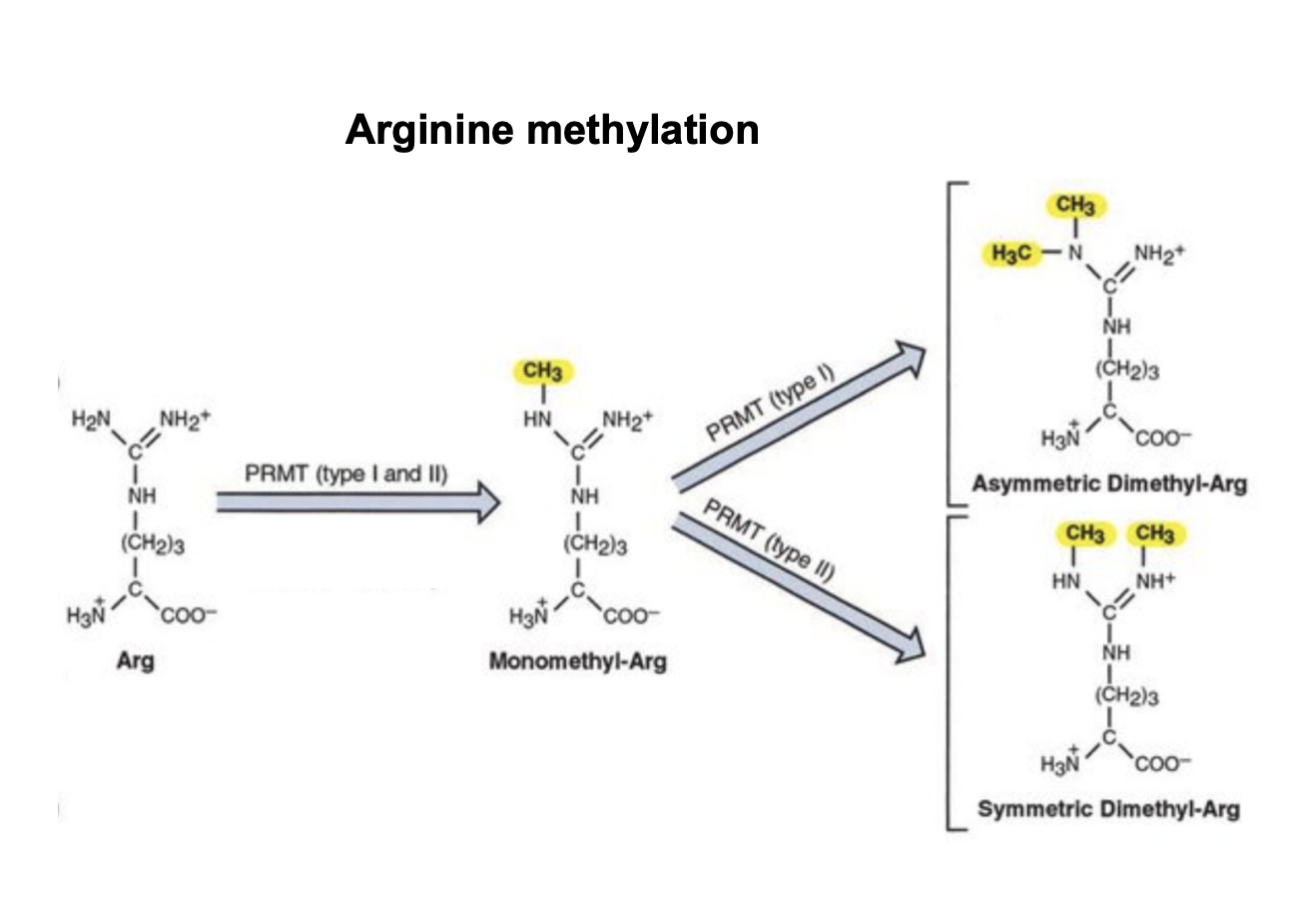
Arginine acetylation and methylation
Acetylation→ lose charge and big
Methylation
Serine phosphorylation
What do these modifications do?
comprise epigenetic information
heritable information that is ot encoded in the DNA sequence
genetic locus is activated/inactivated
under given set of cellular circumstances
What is this concept known as?
histone code
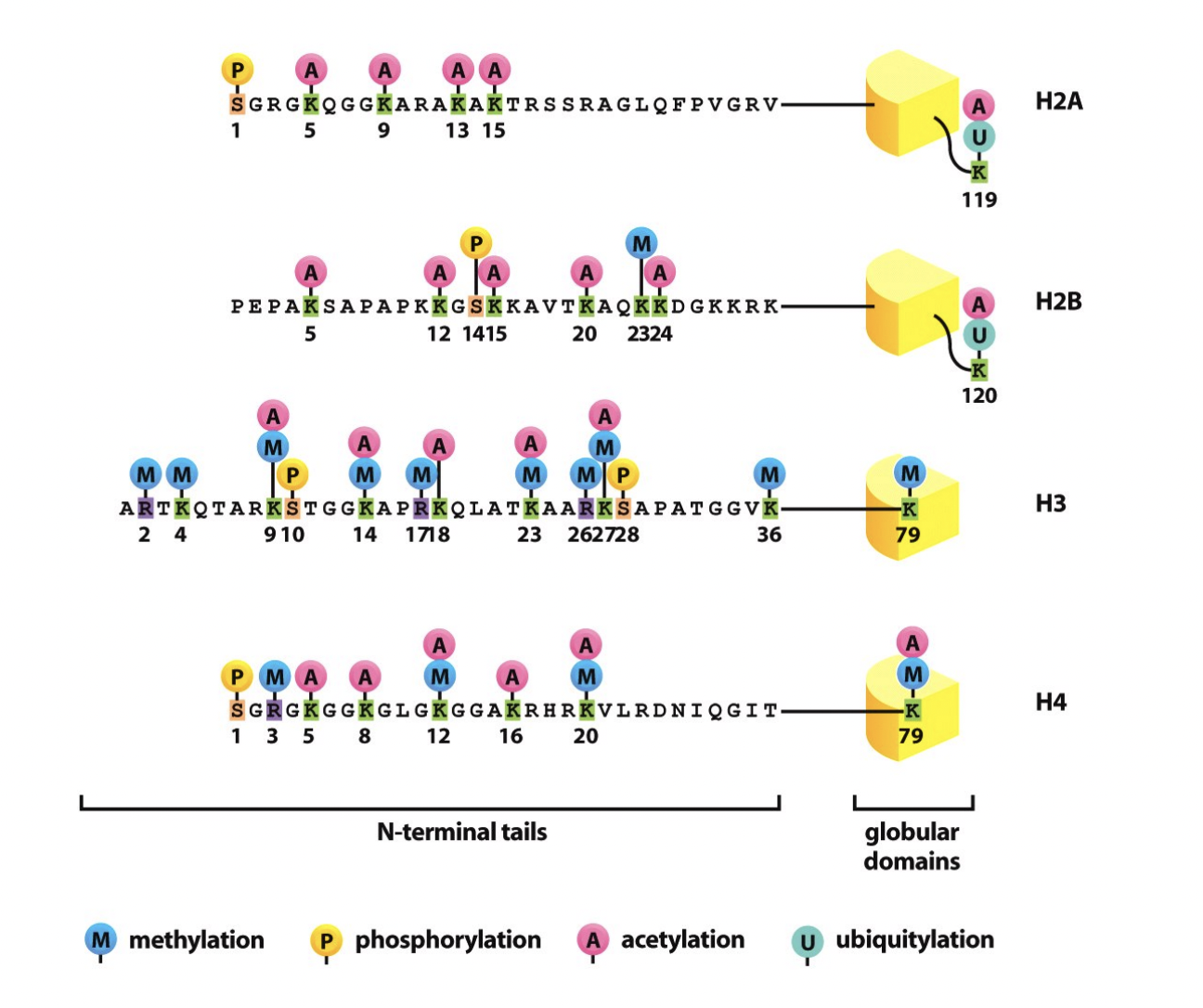
What happens to this hitone code?
‘read’ by proteins that bind specifically to modified histone tailed
e.g of this?
Lysine 9 of histone H3
Acetylated:
Specified active state:
Trimethylated:
Specified inactive state
by attracting different binding proteins
Permenet/reversible?
Sates are reversible
modifications cna be removed and new modification can be added
Therefore the cell’s chromatin is
dynamic entity
regulated changse in its strucutre occur as it
responds to their environment
grow
differentiate