Lecture 14: Movement Across Membranes
1/19
Earn XP
Description and Tags
Morris: Chapter 5, section 5.2
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
Fluidity of biological membranes
Membrane fluidity must be maintained. Lipid composition of membranes can be changed by:
desaturation of fatty acid chains
exchange of fatty acid chains
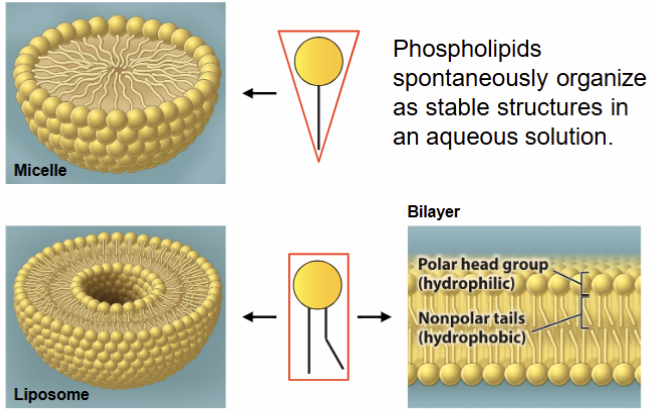
Phospholipid organization
phospholipids spontaneously organize as stable structure in aqueous solution
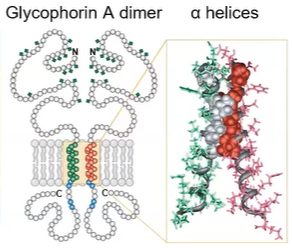
Transmembrane proteins
transmembrane protein domain is a peptide sequence that is largely hydrophobic (uncharged) and spans across the plasma membrane
alpha helix is the most common protein structure element
9 amino acids with hydrophobic side chains
glycine, alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan
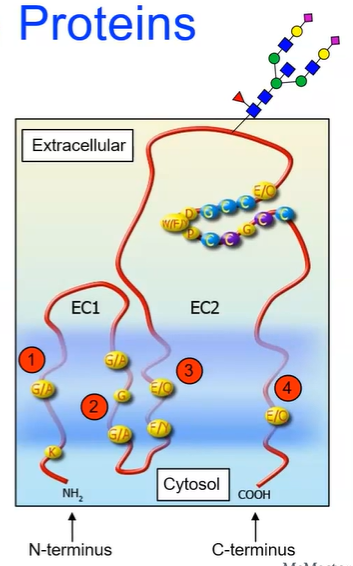
Tetraspanins (TM4SFs)
super family of membrane proteins found in all multicellular eukaryotes
four transmembrane alpha-helices
two extracellular domains
one short (EC1)
one long (EC2)
can be glycosylated (attachment of a carbohydrate molecule) on the long extracellular
plays a role in: cell adhesion, motility, proliferation, etc
Movement across cell membrane
lipid bilayer don’t allow many substances to pass through freely:
can: small, uncharged molecules cross membranes relatively easy (O2, CO2, NO)
can’t: large, polar, charged molecules (Ca+, Glucose, Na+, K+)
can transport with mechanisms/controlled transport
There are 4 basic mechanisms for moving molecules
Passive (1-3), Active (4)
simple diffusion
diffusion through a channel
facilitated diffusion
active transport
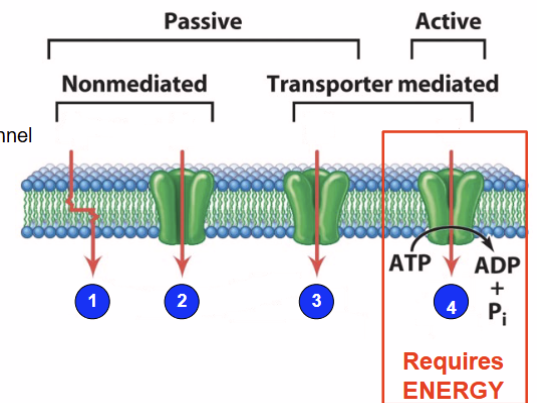
Crossing membrane: passive
relies on molecular concentration, small neutral molecules
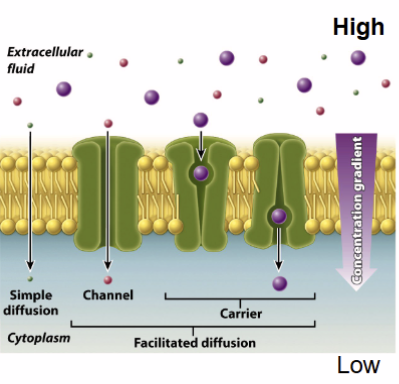
Crossing membrane: simple diffusion
goes down a concentration gradient

Crossing membrane: channels
down concentration gradient, passive, small charged molecules
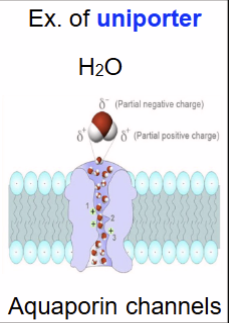
uniporter only 1 molecule, i.e. sodium channel
ion channel: gated open/close in response to different stimuli
ligand-gated channels
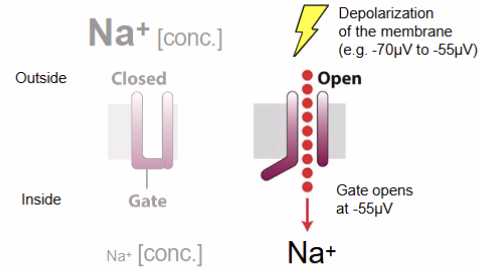
voltage-gated channel
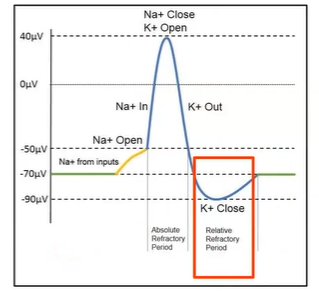
What do voltage-gated channels do?
voltage-gated channel: respond to charge changes across membrane; i.e. Na+ and K+
cell membrane potential: difference in charge across membrane
action potential: passage of electric signal down a nerve axon
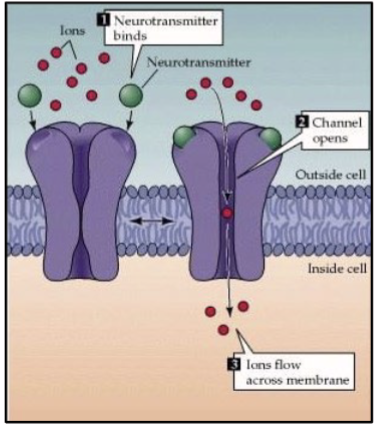
What do ligand-gated channels do?
ligand-gated channels: responds to binding of a specific molecule called a ligand; i.e. acetylcholine receptor
the binding produces a conformational change in the structure of the receptor/channel
Tetrodotoxin (TTX)
potent neurotoxin
discovered in oufferfish
Na+ channel blocker → inhibits the firing of action potentials in neurons by binding to voltage-gated sodium channels in nerve membranes and blocking the passage of Na+ ions into the neuron
prevents nervous system from carrying out messaging
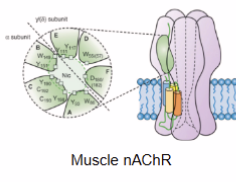
Curare
mixture of organic compounds used as a paralyzing tool
competitive antagonist of the nicotonic acetylcholine receptor (nAChR)
occupies the same position on the receptor as ACh and elicits no response, thus is an example of depolarizing muscle relaxant
Crossing membrane: carriers
facilitated diffusion
glucose transporter: import glucose down a concentration gradient
symporter
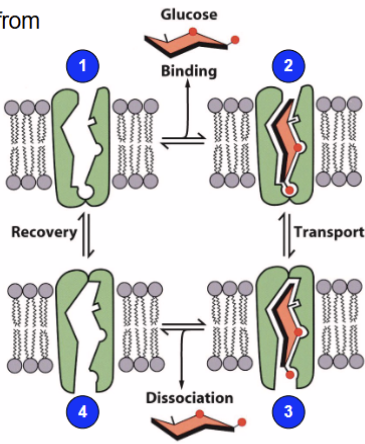
Crossing membrane: carriers - facilitated diffusion
Substrate binds to integral membrane protein called a facilitative transporter, resulting in transporter conformation releasing the compound on other side moving down the concentration gradient
Crossing membrane: carriers - glucose transporter
import glucose from blood down concentration gradient using facilitator
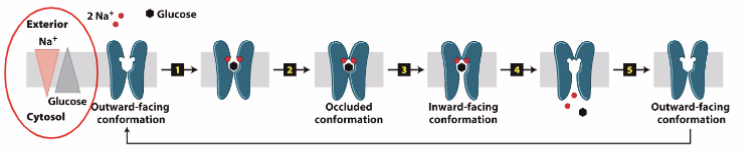
Crossing membrane: carriers - symporter
not reliant on concentration gradient (low → high), relies on chemical gradient of another molecule that wouldn’t reach equilibrium
e.g. Na+ Glucose Symporter: outside, high Na+ low glucose, inside, low Na+ high glucose → 2Na + 1 glucose binds → conformational change (occluded conformation = closed both sides) → inward and releases the three → return to resting position/outward
Crossing membrane: carriers - antiporter
concentration gradient of one molecule is used to transfer second molecule in opposite directions
e.g. sodium-proton exchanger: transports Na+ into cell and protons out of cell
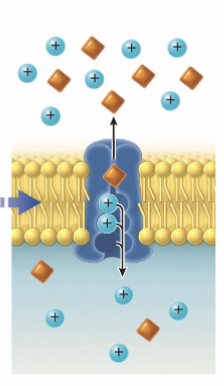
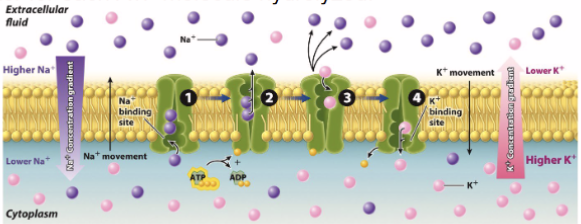
Crossing membrane: active transport
substrate binds to to integral protein/active transporter → hydrolysis of ATP causes conformation to release molecules onto other side of membrane
Na+/K+ pump 3Na leaves and 2K enters and high Na+ concentration outside of cell
maintains cell size