MIC 105 MT 1 lecture 5
1/72
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
73 Terms
Nitrogen
• 3rd-most common
element in most
organisms
• N2 gas = 79% of
atmosphere
• Earth's crust contains N
in ammonium salts, but
is almost entirely
inaccessible to
microbes
The Nitrogen Triangle
• We can simplify
nitrogen cycling into
the conversions
between three (ok, 4)
key molecules
• NO2/NO3: oxidized
forms
• NH4: reduced form
• N2: Oxidation # = 0
most reduced form of N
NH3 and NH4+
Most oxidized form of N
NO3-, HNO3
which type of N is incorporated into cells
NH3 NH4
what is the most abundant form of N
N2
Denitrification
NO3--> N2
Nitrogen fixation
N2 -> NH4+
Nitrification/Comammox
NH4+-> NO3-
Assimilatory Nitrate reduction
NO3--> NH4+
N≡N bond is
very stable
Nitrogen Fixation bacteria
proteobacteria: Azomonas, acidithiobacillus, purple bacteria, rhizobium
firmicutes:Clostridium, Heliobacteria
Cyanobacteria:gloeothece
Archaea:methanosarcina, methanocaldococus
chlorobi: green sulfure bacteria
actinobacteria: frankia
Nitrogenase
key enzyme
complex, reduces N2 to NH3.
• very oxygen sensitive and is
rapidly and irreversibly
inactivated by oxygen.
N2 fixation energy consumption
N2 + 8e- + 8H+ + ~18 ATP -> 2 NH3 + H2 + ~18 ADP + ~20Pi
how many electrons are needed to fix N
8, 6 for the reaction and 2 to make H2 as the product
N2 fixation gene cluster (Klebsiella)
~20 genes are required for nitrogenase production,
homologous in all known organisms.
N2 evolution
N2 fixation evolved once, was likely present in an early ancestor, lost
from some lineages, and acquired by others through horizontal gene
transfer.Anaerobes: e.g., Clostridium, green sulfur (Chlorobi), etc.?
No Oxygen = No Problem!
Nitrogenase production is highly regulated:
only made under
anoxic nitrogen-limiting conditions.
how Microaerophiles deal with O2
intracellular O2 is very low because it is used as fast as it diffuses in.
Respiration rate keeps O2 concentration low enough to protect nitrogenase.
Azotobacter
obligate aerobes capable of N2
fixation under full oxygen atmosphere
High respiration rate, faster than oxygen
diffusion.
Polysaccharide capsule retards O2 diffusion
into the cell
how Cyanobacteria fix N with O2 present
seperates photosynthesis and N2 fixation in two ways: temporally(time)(unicelluar cyanobacteria), Spatially (filamentous cyanobacteria)
Filamentous cyanobacteria
(Nostoc; Anabaena), form heterocysts
heterocysts
nitrogen-fixing cells, terminally
differentiated, depend on the
adjacent cells for
carbohydrate to generate
reductant.
Genes for nitrogenase, glutamine
synthetase (converts glutamate +
ammonia to glutamine), for increased
respiration rate (e.g. cytochrome oxidase)
are derepressed in heterocysts.
Anabaena
• Heterocysts do not fix CO2 (acquire C from adjacent cells)
• Do not produce O2 (PSII is downregulated)
• high respiration rate
• surrounded by a thick cell wall containing glycolipids and polysaccharides that limits
diffusion of O2.
rhizobia
group of symbiotic N2 fixers, Gram- motile rods, mostly members of the alpha Proteobacteria (and a few Betas). They are aerobic heterotrophs.
rhizobia are only capable of N2 fixation when
in symbiotic association with plants. form root nodules on legumes (plants that bear seeds in pods).
Nodule formation involves
both bacterium- and plant-specific gene expression and
cellular differentiation.
which bacteria is soy beans associated with when orming nodules
Bradyrhizobium japonicum
root nodule formation stages
recognition of correct partner and attachment of the bacterium to the root hair
invasion of the rhizobia into the plant root
diffrentiation of plant cells into nodule tissues and bacterial differentiation into nitrogen fixing bacteroids.
how do the plant and bacterium recognize eachother?
plant roots secrete flavonoids into the rhizosphere(layer of soil surrounding roots)
flavonoids
chemoattractants for rhizobia, and induce rhizobia to produce nod factors (determines host microbe specificity)
nod factors
stimulate root hair curling and producrion of a tube through the root hair called the infection thread for rhizobia to travel through, they also stimulate plant cells to divide, fomring nodules
leghemoglobin
red iron containing O2 binding protein produced when the plant and rhizobia are in association
Sym plasmid
Genes involved in nodule
formation and nitrogen
fixation
Nutrient exchange within the nodule
Fixed nitrogen provided to
the plant.
Rhizobium gets
C-source and a protected
environment.
Frankia
Gram+ filamentous actinomycete
symbiosis with Alder trees and other plants.
Alders
'pioneer' trees that can colonize nutrient poor soil, possibly due to associations
with Frankia, which provides fixed N2.
what can frankia do thats similar to azotobacter
fix nitrogen at full oxygen tension and when in symbiosis
how does frankia fix oxygen in O2 environments?
sequester nitrogenase in terminal swellings that have thick walls that retard gas diffusion, protecting nitrogenase from O2
Potential application of N2 fixation
H2 gas production for biofuel
fate of NH4
easily taken up by microbes and plants, and binds well to soil particles even through rain.
how does NH4 feed into the central metabolism
through generation of glutamine
nitrifying bacteria phyla
proteobacteria, nitrospirae
Nitrification
oxidation of ammonia to nitrate, known to be carried out by 2 physiologically distinct groups
ammonia oxidizers
ammonia to nitrite (NO2)
NH3 + 11/2O2 -> NO2- + H2O
eg nitrosococcus
nitrite oxidizers
convert nitrite to nitrate
NO2- + H2O -> NO3- + 2 H+ + 2 e-
eg nitrobacter
how do these oxidizers fix CO2
they are aerobic chemolithoautotrophs that use the calvin cycle to fix CO2
who is a AOB
beta and gamma proteobacteria and the nitrospira lineage
Ammonia monooxygenase
(AMO; membrane bound) converts ammonia to
hydroxylamine, oxygen is substrate.
NH3 + O2 + 2H+ + 2e--> NH2OH + H2O
evolutionarily related to MMO (CH4 -> CH3OH)
Hydroxylamine
oxidoreductase
(periplasmic) catalyzes NH2OH -> NO2-Energy conserved
as PMF
who is a NOB
α, β, γ and δ Proteobacteria and the
Nitrospira lineage
Nitrite oxidoreductase
catalyzes NO2--> NO3-
Energy conserved as PMF
Ammonia-Oxidizing Bacteria and Nitrite-Oxidizing Bacteria
Obligate aerobes
Widespread in soil and water, especially when ammonia is high, e.g., during protein
degradation [ammonification] and in sewage treatment plants
Nitrite, which is toxic and mutagenic, does not accumulate in the environment.
Tight mutualistic symbiosis between AOB and NOB.
Ammonia oxidizing Archaea
Chemolithoautotrophic ammonia oxidizing marine Archaea
First identified by metagenomic sequencing: found 16S gene on same DNA fragment as amo gene
ammonia-oxidizing archeaon example
Nitrosopumilus maritimus part of Thaumarchaeota
Ammonia oxidizing Archaea habitat
ocean and soil, can grow on nh3 concentration 100x lower than AOB
how do AOA fix CO2?
use 3-hydroxypropionate/4-hydroxybutyrate cycle
"Comammox
complete ammonia oxidation to nitrate by single organisms
genome Nitrospira
had genes for ammonia monooxygenase, hydroxylamine oxidoreductase, and nitrite oxidoreductase (comammox)
Nitrification & Comammox importance/ applications
Detrimental for agriculture. Removes usable nitrogen (NH4
+) from agricultural fields and
introduces it into streams and ponds, which causes algal blooms.
NH4
+ binds to negatively charged soil particles and is available for uptake and utilization.
Nitrate (NO3
-; also usable N source) is very water soluble and readily leached from soils in
rainwater runoff.
Result: more fertilizer must be used.
Nitrapyrin, an inhibitor of nitrification (specifically AMO), is added to ammonia fertilizer
on ag fields
Anammox
Anaerobic ammonia oxidation
NH4+ + NO2--> N2
Oxidization of ammonia with nitrite as the
electron acceptor to yield N2 gas under anoxic
conditions
who uses anammox
planctomycetes
Planctomycetes
Unconventional cell envelope: Gram negative, many have
S-layer
Internal compartments, including a membrane-bound structure containing the
nucleoid
aerobic organoheterotrophs
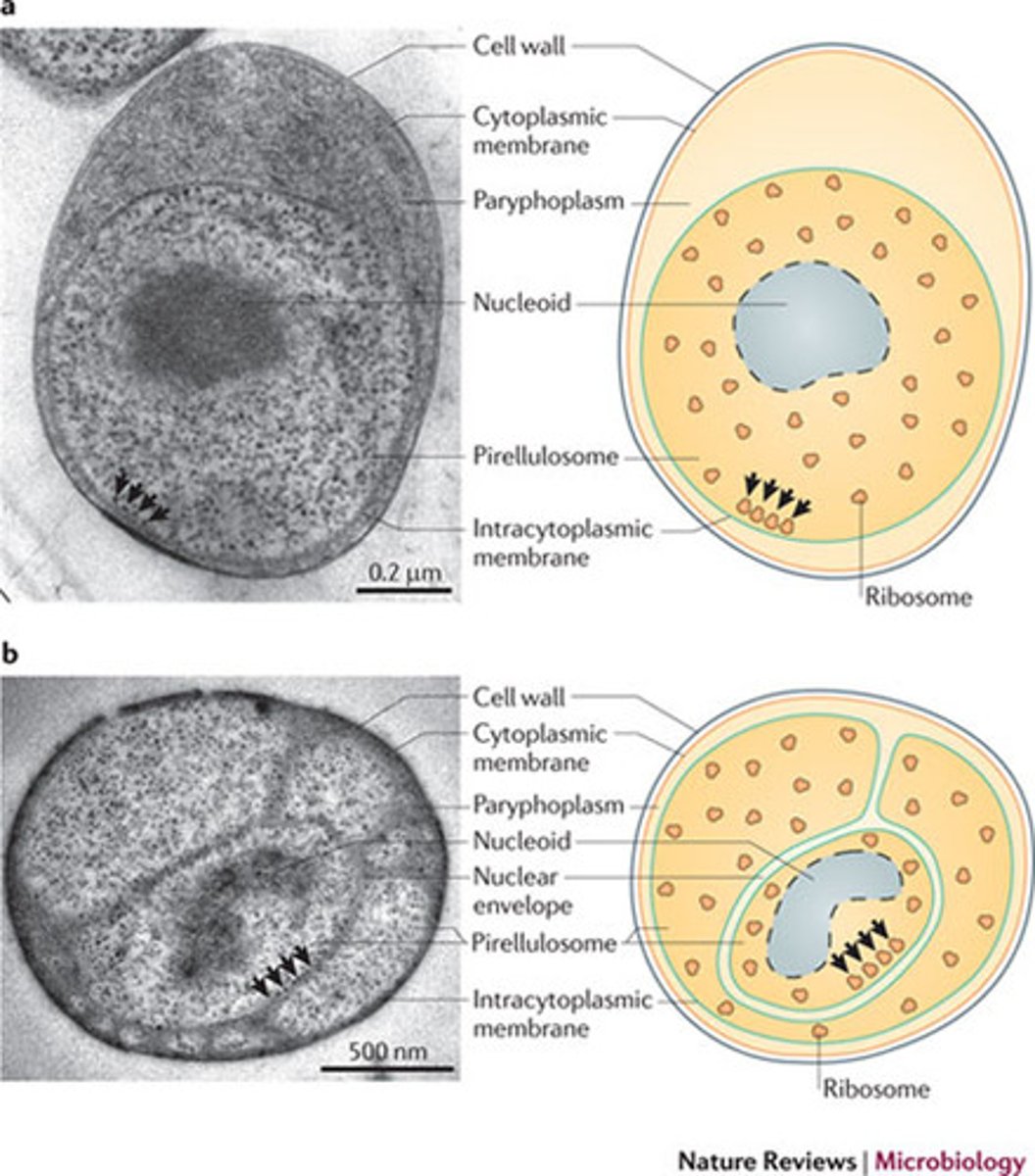
anammox bacteria are
obligately anaerobic
chemolithoautotrophs that fix CO2 using the
reductive acetyl-CoA pathway
anammoxosome
compartment in anammox surrounded by a bilayer membrane
Hydrazine is toxic and mutagenic (DNA damaging);
compartmentalize to protect cellular components; ladderanes keep
permeability low
metabolism is slow (reaction cycles 15 times to fix one CO2
molecule; generation time of 10-30 days!), so any leakage of
metabolites would be significant
ladderanes
Unique lipids with ring structures in anammoxosome membrane
reduce membrane
permeability
AAO
NH4+ + NO2--> N2 + 2 H2O
Toxic hydrazine (N2H4) is an intermediate
Key enzymes of AAO
NIR - Nitrite reductase
HZS - hydrazine synthase
HDH - hydrazine dehydrogenase
Denitrification
NO3--> NO2--> NO -> N2O -> N2
conversion of nitrate to gaseous nitrogen compounds
nitrate respiration
Bacteria carrying out have membrane-bound or periplasmic enzymes to
couple substrate oxidation to nitrate reduction to generate PMF, which allows ATP
synthesis.
Key enzyme of nitrate respiration
itrate reductase: NO3--> NO2-
Genes encoding nitrate reductase are only expressed anaerobically, as O2 is the preferred
terminal electron acceptor for respiration
Applications/ importance of Denitrification
Detrimental for agriculture: converts NO3
-, a good nitrogen source, into nitrogen gas.
Nitrogen oxides (NO, N2O) = greenhouse gases and contribute to global warming.
Good for wastewater treatment:
Nitrates linked to reproductive
problems, cancer, and 'blue
baby syndrome', in which nitrite
decreases the O2 carrying
capacity of hemoglobin.
Denitrification & Anammox importance
Both are beneficial in wastewater treatment.
Remove usable sources of nitrogen, so water released into rivers and streams has fewer
algal blooms, reduction in anaerobic 'dead zones' which result in fish death.