Nonenzymatic Protein Function and Protein Analysis
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
33 Terms
What happens to an enzyme after it catalyzes a reaction?
”Enzymes = Everlasting workers"
Enzymes are not consumed or permanently changed by the reactions they catalyze.
They can be reused many times to catalyze the same reaction.
What do lyases do?
Lyases are enzymes that break bonds without using water or redox reactions.
They often form a new double bond or add groups to double bonds.
What do isomerases do?
Internal Shuffle" — same parts, new arrangement.
Isomerases are enzymes that rearrange atoms within a molecule to form an isomer (same molecular formula, different structure).
They do not add or remove atoms — just rearrange the existing ones.
What do ligases do?
Ligases are enzymes that join two molecules together, often using energy from ATP hydrolysis.
They form new covalent bonds (C–C, C–O, C–N, or C–S).
What do hydrolases do?
"Hydrolases = Hydrolyze with H₂O"
Hydrolases are enzymes that break bonds by adding water (hydrolysis).
They split large molecules into smaller ones.
What do transferases do?
Transferases are enzymes that transfer a functional group (like phosphate, methyl, or amino) from one molecule (donor) to another (acceptor).
What is an exergonic reaction?
An exergonic reaction is a chemical reaction that releases free energy (ΔG < 0).
It is spontaneous — meaning it can proceed without an input of energy.
"Exergonic = Energy exits" — energy flows out, reaction goes forward.
How do enzymes affect ΔG, ΔH, and reaction equilibrium?
"Enzymes change the clock, not the destination"
Enzymes do not change:
ΔG (free energy change)
ΔH (enthalpy change)
The position of equilibrium
Enzymes do:
Lower the activation energy
Speed up reaction rate (kinetics)
Allow equilibrium to be reached faster
How do enzymes catalyze reactions?
"Stabilize, Surround, Stick"
Enzymes speed up reactions by:
Stabilizing the transition state → lowers activation energy
Providing a favorable microenvironment (e.g., pH, polarity)
Forming temporary bonds with the substrate (induced fit, covalent catalysis)
What is the active site of an enzyme?
The active site is the specific region on an enzyme where the substrate binds and catalysis occurs.
It has a unique shape and chemical environment tailored to the substrate.
How is binding to the enzyme active site explained?
"Lock & Key = perfect fit; Induced Fit = enzyme hugs substrate"
Two models explain substrate binding:
Lock and Key Theory:
The substrate fits exactly into the enzyme’s active site like a key in a lock.
Induced Fit Model:
The enzyme changes shape upon substrate binding to fit the substrate more snugly, enhancing catalysis.
What do some enzymes require to be active?
"Cofactors and Coenzymes = Enzyme’s sidekicks" — they help enzymes perform their job!
Cofactors: Usually metal cations (e.g., Mg²⁺, Zn²⁺) that help stabilize enzyme structure or participate in catalysis.
Coenzymes: Small organic molecules (e.g., NAD⁺, FAD, vitamins) that assist in electron or group transfer.
What happens to enzyme reaction rate as substrate concentration increases?
As substrate concentration increases, the reaction rate increases until it reaches a maximum velocity (Vmax) where all enzyme active sites are saturated. Beyond this point, adding more substrate does not increase the rate.
"More substrate, faster rate — until full, then steady state."
How do Michaelis-Menten and Lineweaver-Burk plots represent enzyme kinetics?
Michaelis-Menten plot: Shows reaction rate (v) vs. substrate concentration [S] as a hyperbola approaching Vmax.
Lineweaver-Burk plot: A double reciprocal plot (1/v vs. 1/[S]) that linearizes the Michaelis-Menten curve, making it easier to determine Vmax and Km.
How can enzymes be compared using Km and Vmax?
Km (Michaelis constant): Indicates the substrate concentration at half Vmax.
Lower Km = higher affinity for substrate.
Vmax: The maximum reaction velocity an enzyme can achieve at saturating substrate.
Comparing enzymes:
Higher affinity = lower Km
Faster max rate = higher Vmax
Why do cooperative enzymes display a sigmoidal curve in substrate binding?
Cooperative enzymes have multiple binding sites, and binding of substrate to one site increases the activity or affinity at other sites (positive cooperativity). This causes a sigmoidal (S-shaped) curve when plotting reaction rate vs. substrate concentration.
"Cooperative enzymes: one binds, others jump in — making the curve S-shape!"
What kind of curve does cooperative enzymes display?
A sigmoidal curve
Why do cooperative enzymes display a sigmoidal curve, and what does it mean?
Why:
Cooperative enzymes have multiple subunits and binding sites. When one substrate binds, it increases the affinityof the remaining sites for more substrate (positive cooperativity).What it means:
Low [S] (first flat region): Low activity because few sites are occupied.
Middle steep slope: Small increases in [S] cause large increases in activity due to cooperative binding.
High [S] (top plateau): Enzyme is saturated — all sites filled, rate at Vmax.
Q: How does increasing solute concentration affect proteins?
in vivo
Significant deviations can denature the enzyme, causing:
Loss of secondary structure (α-helices, β-sheets)
Loss of tertiary structure (overall 3D fold)
Loss of quaternary structure (if multiple subunits are present)
Denaturation destroys the active site, leading to loss of catalytic activity.
salinity impacts enzymes in vitro or in vivo?
In vitro
How are enzyme pathways regulated?
Enzyme pathways are highly regulated to control metabolic flow and maintain homeostasis.
They are subject to:
Inhibition (slowing or stopping activity)
Activation (increasing activity)
The main types of inhibition:
reversible inhibition: Inhibition can be overcome by removing the inhibitor using mild laboratory treatment or using a compound with a greater affinity.
competitive inhibition
non competitive inhibition
mixed inhibition
uncompetitive inhibition
irreversible inhibition: The enzyme’s function cannot be restored without making new enzyme molecules. "Irreversible = locked forever" — once bound, the enzyme is done.
What is competitive inhibition and how does it affect Km and Vmax?
Definition: Inhibitor competes with the substrate for binding at the active site.
Effect:
Km increases (more substrate needed to reach half Vmax).
Vmax stays the same (can be reached if enough substrate is added).
Overcome by: Increasing substrate concentration.
What is noncompetitive inhibition and how does it affect Km and Vmax?
Definition: Inhibitor binds to an allosteric site on the enzyme, not the active site, regardless of substrate binding.
Effect:
Vmax decreases (enzyme’s maximal activity is reduced).
Km stays the same (substrate affinity is unchanged).
Cannot be overcome by adding more substrate.
What is uncompetitive inhibition and how does it affect Km and Vmax?
Definition: Inhibitor binds only to the enzyme–substrate complex, not the free enzyme. This is to an allosteric site on the ES complex
Effect:
Vmax decreases (less enzyme activity overall).
Km decreases (enzyme–substrate affinity appears higher).
What is mixed inhibition and how does it affect Km and Vmax?
Definition: Inhibitor can bind to either the active site on the free enzyme or the allosteric site on the enzyme–substrate complex, but with different affinities.
Effect:
Vmax decreases (overall enzyme activity drops).
Km increases or decreases depending on whether the inhibitor prefers free enzyme (Km ↑) or ES complex (Km ↓).
The main types of activation:
allosteric sites
phosphorylation
glycosylation
zygomens
What can binding at an allosteric site do to an enzyme?
Allosteric sites are distinct from the active site.
Binding of molecules at these sites can modulate enzyme activity by:
Increasing substrate affinity (makes enzyme bind substrate more tightly)
Increasing enzymatic turnover (makes enzyme convert substrate to product faster)
Allosteric regulation can also decrease these properties if the effector is an inhibitor.
What post-translational modifications can affect enzymes?
Phosphorylation: covalent modfication with a phosphate
Glycosylation: covalent modification with a carbohydrate
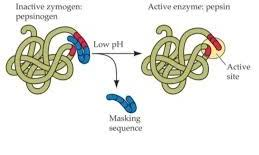
Zymogens:
Zymogens are inactive precursors of enzymes.
They require a specific cleavage to become active enzymes.
These type of proteins compose the cytoskeleton, anchoring protiens, and much of the extracellular matrix:
structural proteins
The most common type of structural proteins:
Collagen
Keratin
Elastin
Actin & Tubulin
What is the shape of structural proteins?
They are typically fibrous in nature — long, thin, and rope-like.
Fibrous structure gives them tensile strength and stability.