Cell Cycle 2
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
49 Terms
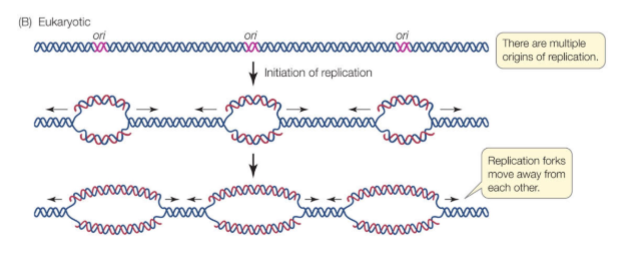
True/False DNA Replication initiates from multiple origins
True
What decides where DNA replication initiates in yeast & higher eukaryotes?
Yeast: origins defined by specific DNA sequences.
Higher eukaryotes: specified by binding of the Origin Recognition Complex (ORC).
What are the two main steps of DNA replication?
Initiation – activation of an origin, giving rise to two replication forks.
Elongation – polymerization of new DNA strands.
What are the three defining keywords of DNA replication?
Bidirectional – replication proceeds in both directions from the origin.
Semi-conservative – each daughter DNA contains one parental strand and one new strand.
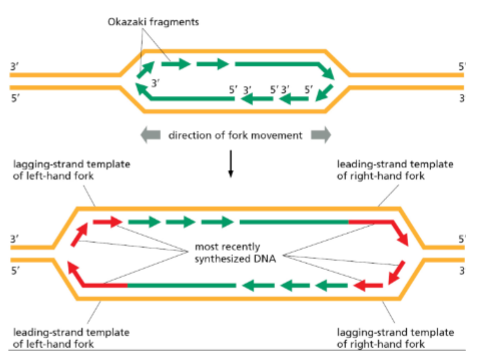
Semi-discontinuous – continuous synthesis on the leading strand, discontinuous on the lagging strand.
Why are multiple origins required for large genomes like humans
Because replication forks duplicate DNA at ~2 kb/min. If humans had only one origin, replication would take ~1000 days. Multiple origins reduce this to hours.
Why do cells have to make sure that origins are activated only once per cell cycle?
Both under-activation and over-activation cause genome instability.
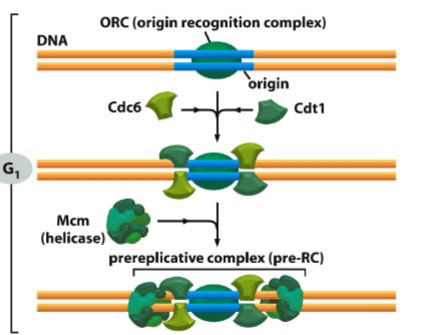
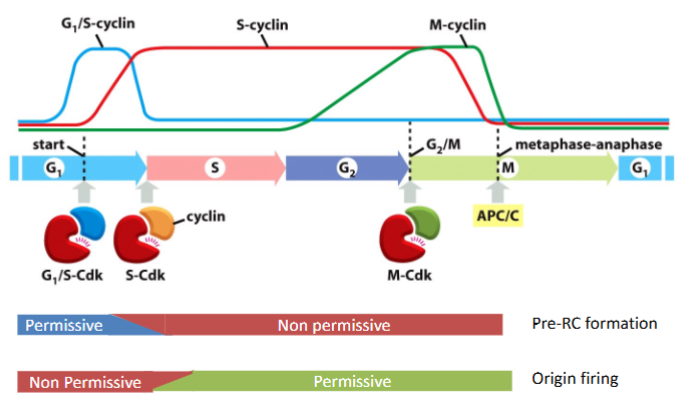
What is the pre-Replicative Complex (pre-RC) and when does it form?
The pre-RC is a protein complex that licenses DNA for replication. It forms during G1 phase at replication origins
What processes of origin activation and disassembly of pre-RC take place in the S phase
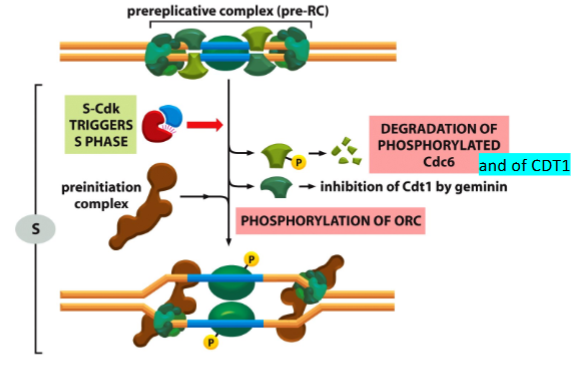
What processes of origin activation and disassembly of pre-RC take place in the G2/M phase
Completion of DNA replication
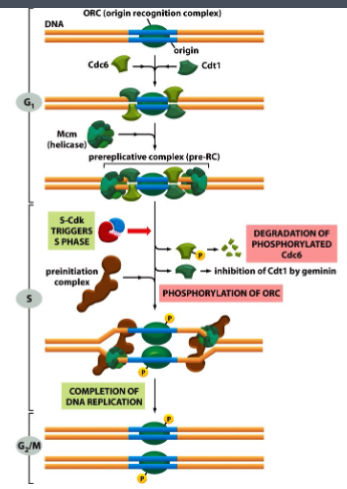
What is CDT1 and what role does it play in DNA replication?
CDT1 is a licensing factor that loads MCM helicase onto origins during G1. It is essential for pre-RC formation.
What happens to CDT1 after replication initiation?
CDT1 is removed/degraded to prevent re-replication within the same cell cycle.
What triggers the S phase
S-Cdk
What is Geminin and how does it regulate DNA replication?
Geminin binds and inhibits CDT1, blocking MCM loading during S-phase and G2, ensuring that origins are not reused.
Study this
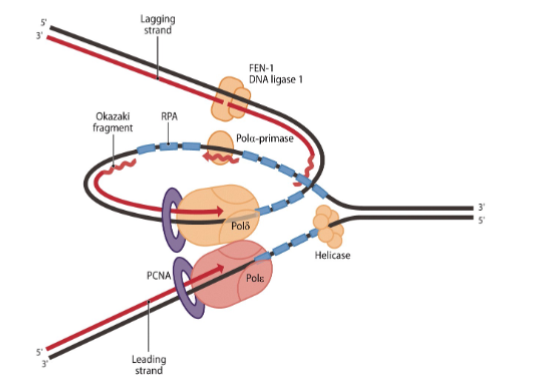
Which three DNA polymerases function at each replication fork?
DNA polymerase α (alpha): Initiates DNA synthesis by adding primers.
DNA polymerase δ (delta): Extends Okazaki fragments on the lagging strand.
DNA polymerase ε (epsilon): Main polymerase on the leading strand.
What do DNA polymerases require to initiate DNA synthesis?
A free 3′-OH group, provided by an RNA primer.
In which direction do DNA polymerases synthesize DNA?
Only in the 5′ → 3′ direction. (DNA polymerases can add nucleotides only to the 3’ end of a DNA strand)
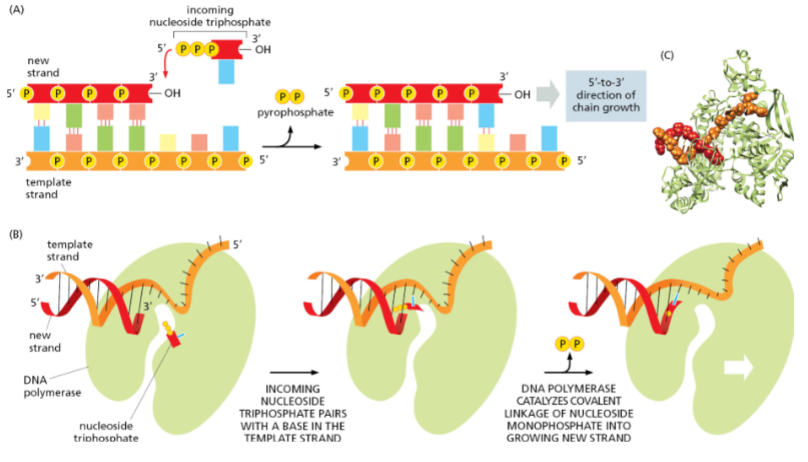
Why is the lagging strand synthesized discontinuously?
Because DNA polymerases can only extend 5′ → 3′, so lagging strand synthesis occurs in fragments (Okazaki fragments) opposite the replication fork’s movement.
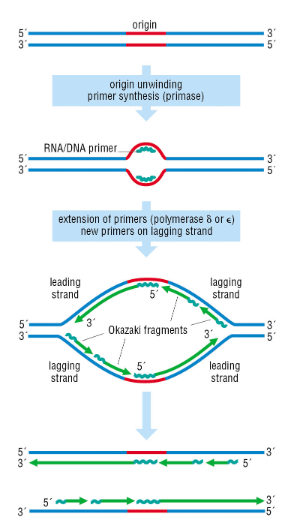
What problem arises at the ends of linear chromosomes & how is it solved
The end replication problem — inability to fully replicate chromosome ends. This is solved by telomerase
What are the key steps in lagging strand DNA synthesis
a. Helicase unwinds DNA, RPA (Replication Protein A) stabilizes single strands.
b. DNA polymerase α–primase synthesizes a short RNA-DNA primer.
c. DNA polymerase α is displaced and replaced by polymerase δ.
d. DNA polymerase δ/ε extend the primer.
e. Downstream RNA primers are removed by nucleases.
f. Okazaki fragments are ligated by DNA ligase
Which polymerase is the main enzyme on the leading strand?
DNA polymerase ε.
What mechanism ensures high fidelity in DNA replication?
The proofreading activity of DNA polymerases δ and ε.
What type of enzymatic activity is used for proofreading?
3′ → 5′ exonuclease activity, which removes incorrectly paired nucleotides from the newly synthesized strand.
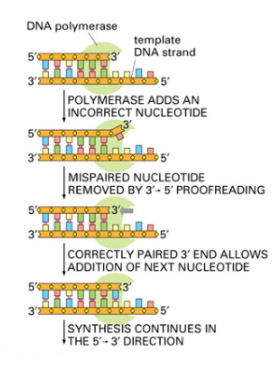
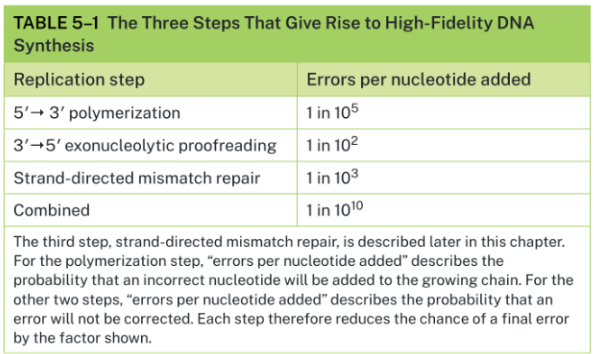
Name the 3 steps that give rise to high-fidelity DNA Synthesis
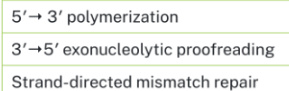
Which DNA polymerases are commonly mutated in cancer?
DNA polymerase δ (Pol δ) and DNA polymerase ε (Pol ε).
Where do cancer mutations usually occur?
In the proofreading exonuclease domains
What is a centrosome
The major microtubule-organizing center (MTOC) of the cell
Centrosome structure
2 centrioles (barrels of microtubules composed of tubulin and centrin) surrounded by cloud of pericentriolar material (PCM).
PCM contains g-tubulin ring complex that nucleates microtubules.
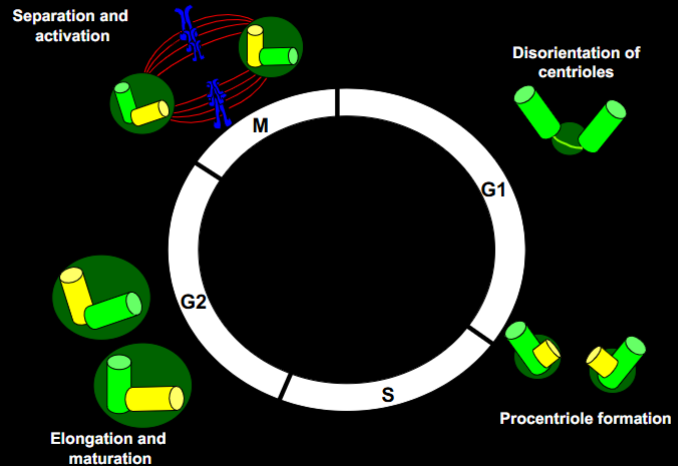
What are the key steps in centrosome duplication?
G1: Centrioles disoriented.
S phase: Procentriole formation begins next to each parental centriole.
G2: Elongation and maturation of procentrioles.
M phase: Separation and activation of centrosomes to form spindle poles.
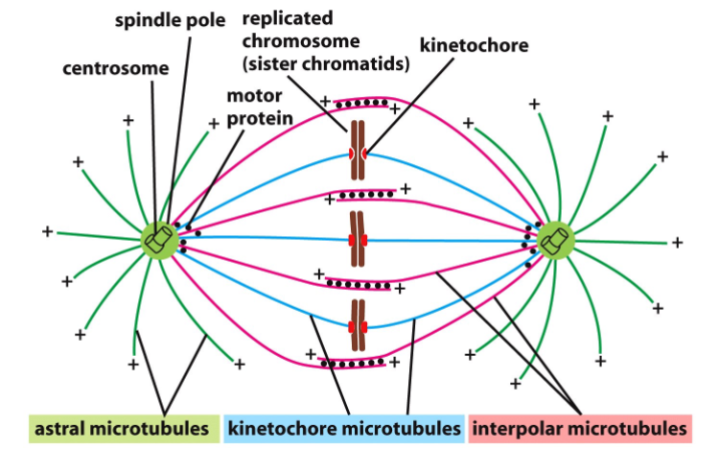
Structure of centrosomes when attached to sister chromatids
How do checkpoints usually regulate progression?
Through negative feedback — stopping progression if errors are detected, rather than accelerating positive signals.
When does the G1 checkpoint occur?
At the end of G1, just before S-phase.
What decisions can a cell make at the G1 checkpoint?
Proceed to divide.
Delay division.
Exit the cycle into G0 (quiescence - arrested stage)
Which cyclins and CDKs are involved in passing the G1 checkpoint?
Cyclin D with CDK4/6 phosphorylate retinoblastoma protein (Rb).
Cyclin E with CDK2 further phosphorylate Rb.
How does Rb phosphorylation affect the cell?
Activates E2F transcription factor, which promotes expression of S-phase genes.
What is the role of p53 at the G1 checkpoint?
tumor suppressor protein
p53 halts the cell cycle if DNA damage is detected, preventing entry into S-phase
What happens when p53 is mutated
Loss of ability to activate target genes like p21.
p21 normally inhibits G1 cyclin-CDK complexes.
Without this inhibition, damaged cells enter S-phase.
How common are p53 mutations in human cancers?
Found in >50% of cases.
What prevents cells from entering mitosis if replication is incomplete?
A DNA replication stress checkpoint.
What activates the DNA replication stress response
Accumulation of single-stranded DNA (ssDNA) at stalled replication forks.
Which types of proteins are recruited to stalled replication forks?
Replication proteins and signaling molecules
Which kinases amplify the stress signal?
ATR and CHK1 kinases.
TR and CHK1 kinases amplifying the stress signal leads to what
Degradation of Cdc25 phosphatase.
This inhibits CDK1.
Cell cycle arrest occurs until replication stress is resolved.
What major transition occurs after successful DNA replication and checkpoint clearance?
Entry into mitosis for chromosome segregation.
What does the spindle-attachment checkpoint (SAC) monitor?
Ensures sister chromatids are correctly attached to the mitotic spindle before anaphase.
What structure is monitored by the SAC?
The kinetochore (protein complex at centromeres where spindle fibers attach).
What happens if kinetochores are unattached?
They send a negative signal to inhibit Cdc20–APC (anaphase-promoting complex).
This delays anaphase until proper attachment occurs.
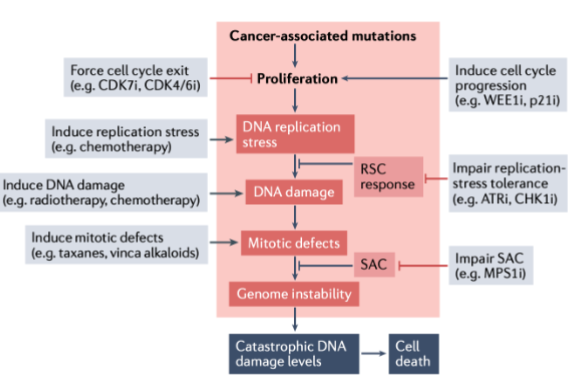
How do cancer associated mutations lead to cell death